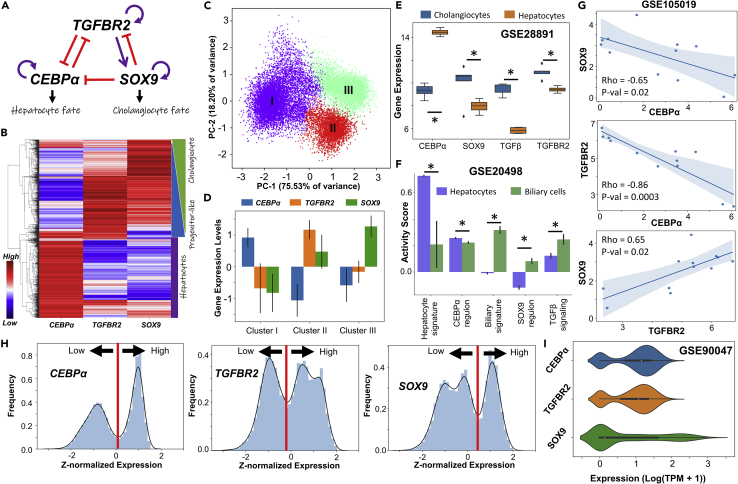
Figure 1.
Multiple phenotypes enabled by a regulatory network driving cell-fate decisions in liver
(A) Gene regulatory network (GRN) underlying plasticity in the hepatocyte-cholangiocyte cell-fate decision. Purple arrows represent activation links; red hammers represent inhibitory links.
(B) Heatmap showing hierarchical clustering in ensemble of steady state solutions obtained from the GRN. Red represents higher expression levels while blue denotes lower expression levels, plotted as z-scores.
(C) Principal Component Analysis (PCA) showing three major clusters in steady state solutions obtained.
(D) Quantification of steady state levels from the three clusters identified by PCA. Error bars represent the SD over n = 3 replicates.
(E and F) (E) Gene expression levels of SOX9, c/EBPα, TGFβ and TGFBR2 in hepatocytes and cholangiocytes (GSE28891).
(F ) Activity levels of the gene expression signatures quantified through single sample Gene Set Enrichment Analysis (ssGSEA) in hepatocytes and biliary cells (GSE20498): c/EBPαregulon, SOX9 regulon and TGFβ signaling. In E, F panels; ∗ represents a statistically significant difference in the activity/expression levels (Students’ two tailed t test; pvalue<= 0.05). (G)Scatterplots showing pairwise correlations in human liver progenitor-like population in culture (GSE105019). Spearman correlation coefficient (Rho) and pvalue (p-val) are given.
(H) Simulation data indicating the multimodal nature of steady state values in the expression of the various nodes in the network.
(I) Single-cell RNA-seq data (GSE90047) for a population composed of hepatocytes, hepatoblasts and cholangiocytes: distributions of gene expression levels of SOX9, c/EBPα, and TGFBR2.