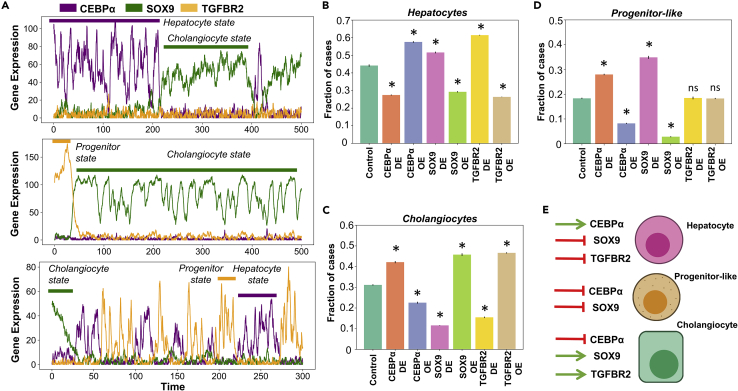
Figure 4.
Stochastic cell state switching and predicted cellular reprogramming strategies
(A) Noise induced stochastic switching simulations of the gene regulatory network (GRN) under different representative parameter sets. Biological states have been defined based on the expression levels of the individual genes. (B-D) Simulation results quantifying the levels of (B) hepatocytes (C) cholangiocytes and (D) progenitor-like cells under various perturbations to the gene regulatory network.
(E) Schematic showing the list of perturbations that are likely to be the most effective in enriching for a given cell type. ∗ represents a statistically significant difference in the proportion of cases resulting in a particular phenotype compared to control case (Students’ two-tailed t test; pvalue<0.05); ns denotes a non-significant difference.