Figure 3. RHBDL4 cleaves distinct soluble ERAD-L substrates.
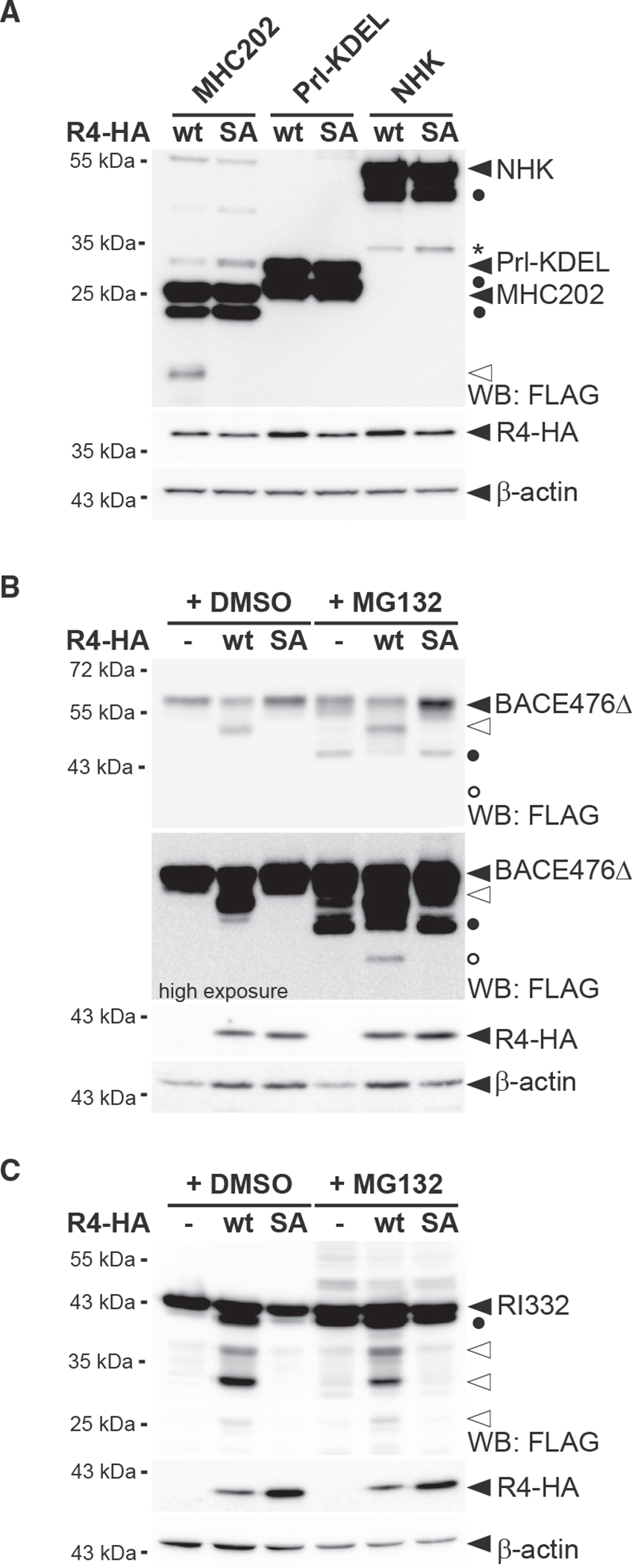
(A) RHBDL4 does not cleave Prl-KDEL and NHK. Hek292T cells were co-transfected with N-terminally FLAG-tagged MHC202, Prl-KDEL, or NHK with either HA-tagged RHBDL4 (R4-HA) WT or the SA mutant and analyzed by western blotting (WB). Filled triangle, full-length glycosylated proteins; open triangle, N-terminal MHC202 cleavage fragment; asterisk, RHBDL4 independent NHK degradation intermediate; filled circle, deglycosylated full-length proteins.
(B) R4-HA WT generates an N-terminal 40-kDa BACE476Δ cleavage fragment (open triangle) that is degraded by the proteasome as shown by increased steady-state level upon MG132 treatment compared with vehicle control (DMSO). Upon proteasome inhibition, the 34-kDa deglycosylated full-length BACE476Δ (filled circle) and traces of a deglycosylated form of the RHBDL4-generated cleavage fragment (open circle) become visible.
(C) Cleavage assay as in (B), but with N-terminally FLAG-tagged RI332 as substrate showing cleavage fragments in the range of 25–35 kDa (open triangles). Filled circle, deglycosylated full-length RI332.
For (A)–(C), representative experiments of three biological replicates are shown and β-actin was used as a loading control.
