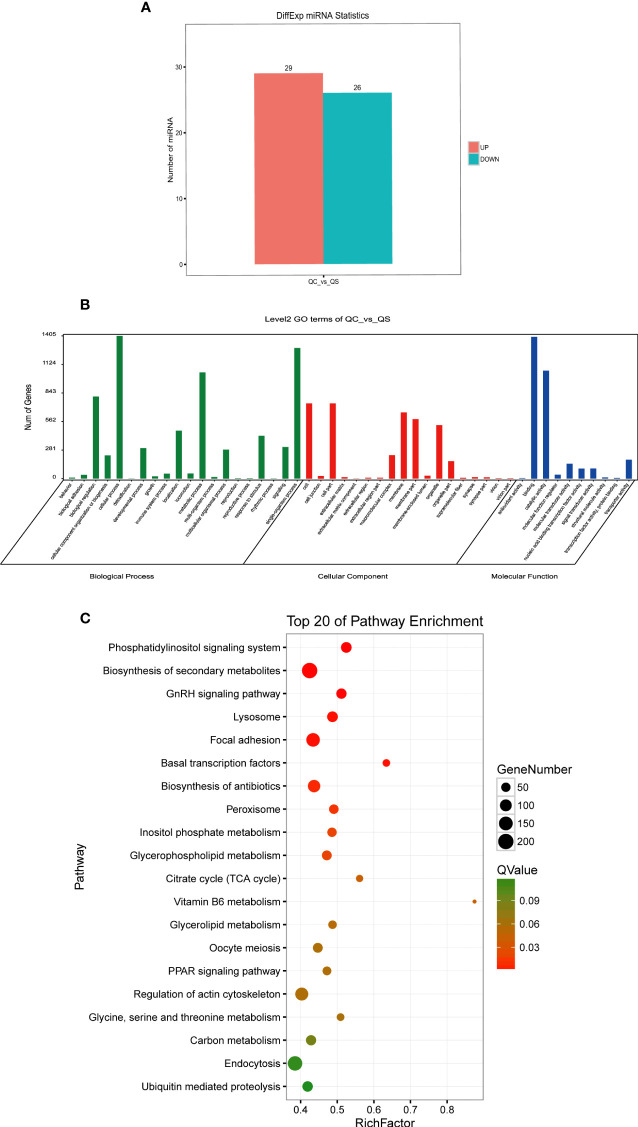
Figure 4.
Identification and functional annotation of differentially expressed miRNAs. (A) Histogram shows differentially expressed miRNAs. Blue and pink indicate decreased and increased expression, respectively. (B) GO enrichment analysis of predicted target genes of DEMs. The abscissa indicates 3 ontologies (molecular function, cellular component and biological process) with 48 GO terms, the vertical coordinate indicates the genes count. (C) KEGG pathway analysis of target genes of DEMs. The redder bubble indicates more obvious enrichment, with smaller Q-value. Size of the point refers to the number of genes within each pathway. DEMs, differentially expressed miRNAs; DEGs, differentially expressed genes; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.