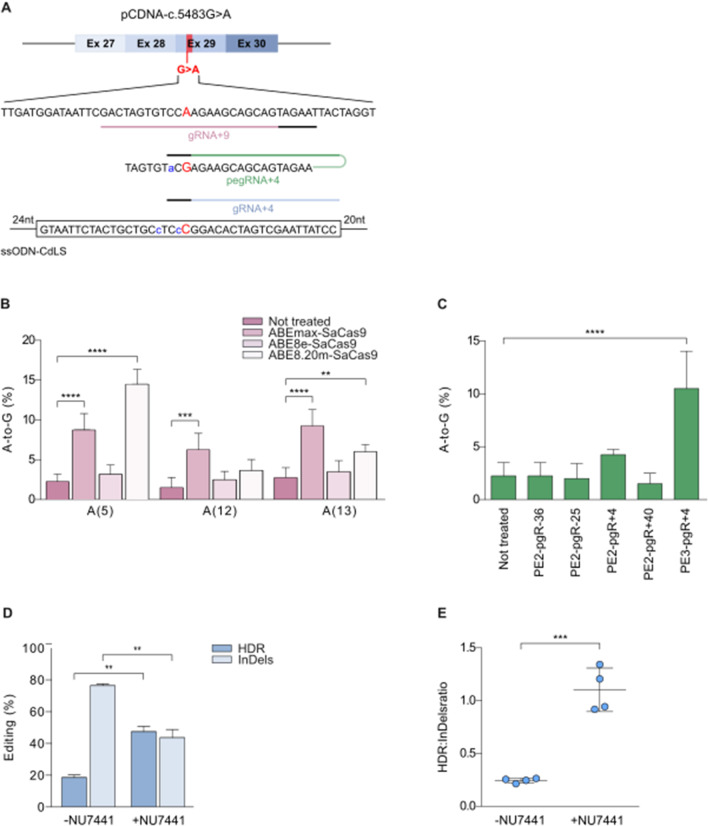
Fig. 1.
Correction of the NIPBL c.5483G > A substitution in a HEK293-CdLS cell model. A Scheme of the NIPBL cDNA (pCDNA–c.5483G > A) used to test the genome editing strategies for the G > A substitution (highlighted in red) in exon 29. The lower panel shows part of the nucleotide sequence and the target gRNA + 4, gRNA + 9 and pegRNA + 4 positions (underlined in pink, green and light blue, respectively, with the PAMs in black) and the ssODN-CdLS sequence (the corrected nucleotide is shown in red, silent mutations are shown in blue). B A-to-G transitions tested in HEK/CdLS clones (cl1 and 2) treated with ABEmax-SaCas9, ABE8e-SaCas9 and ABE8.20 m-SaCas9 combined with gRNA + 9. Modification of the adenines in the targeting window are reported numbered relative to the 5’ distal end of the gRNA, as reported by Rees et al. [17], A(5), A(12) and A(13). n ≥ 4 replicates. Data are means ± SD. Statistical analysis was performed using two-way ANOVA; **P ≤ 0.01, ***P ≤ 0.001, ****P ≤ 0.0001. C A-to-G transitions mediated by PE2 and PE3 strategies using the indicated pegRNAs (pgR + -25, pgRNA-36; pgRNA + 4; pgRA + 40) in HEK293/CdLS clones (cl1 and 2) through plasmid delivery; n ≥ 2 replicates. Data are means ± SD. Statistical analysis was performed using two-way ANOVA; ****P ≤ 0.0001. D Editing efficiencies analyzed by TIDER in HEK293WT–CdLS cells (HEK293/CdLS-cl1) electroporated with SpHiFiCas9-gRNA + 4 RNPs and ssODN-CdLS untreated or treated with DN-PK inhibitor NU7441. E HDR/InDels ratio analyzed in cells treated as in E. Data were obtained from n = 4 experiments. Data are means ± SD. Statistical analysis was performed using two-way ANOVA; **P ≤ 0.01, ***P ≤ 0.001