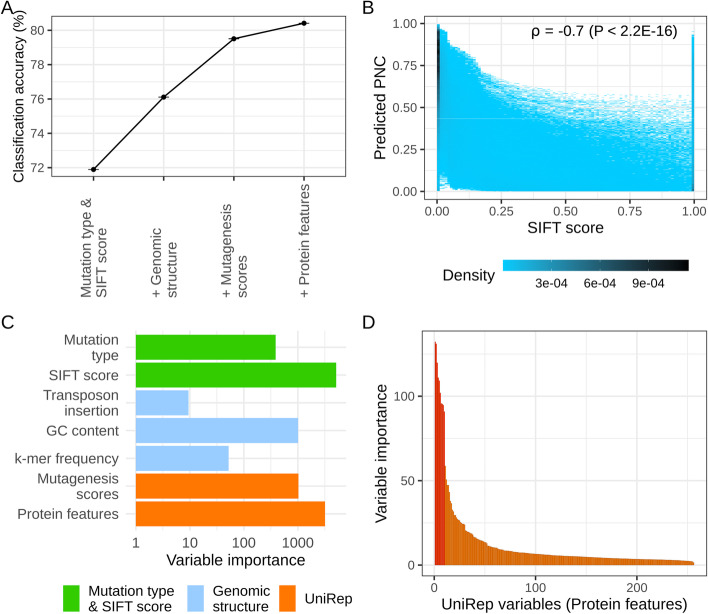
Fig. 3.
Contribution of genomic annotations to prediction accuracy in probability random forests. A Classification accuracy of probability random forests for predicted phylogenetic nucleotide conservation (PNC). Accuracy: percentage of correct calls, i.e., the percentage of sites in chromosome 8 for which predicted PNC (rounded) equaled observed PNC, over three replicates. Accuracy was weighted to account for imbalance with respect to PNC (see “Methods”). Sets of genomic annotations were sequentially added to the set of predictors in probability random forests. Mutation type & SIFT score: Mutation type (missense, STOP gain or STOP loss), SIFT score (with missing values set to 1), and SIFT class (“constrained” if SIFT score ≤ 0.05, “tolerated” otherwise). Genomic structure: GC content, k-mer frequency and transposon insertion. Mutagenesis scores: in silico mutagenesis scores for UniRep variables. Protein features: UniRep variables, generated by the 256-unit UniRep model. B Relationship between SIFT scores and predicted PNC at maize SNPs (observed polymorphisms in Hapmap 3.2.1, a representative panel of inbred lines in maize [15]). Predicted PNC is computed by the full PICNC model, including all genomic annotations. Darker colors indicate higher density of SNPs. ρ: Spearman correlation coefficient. C Variable importance of genomic annotations. Variable importance: corrected impurity measure in probability random forests [18]. D Variable importance of protein features (UniRep variables), ordered in decreasing order. A subset of 10 UniRep variables stood out as contributing most to the prediction accuracy for PNC