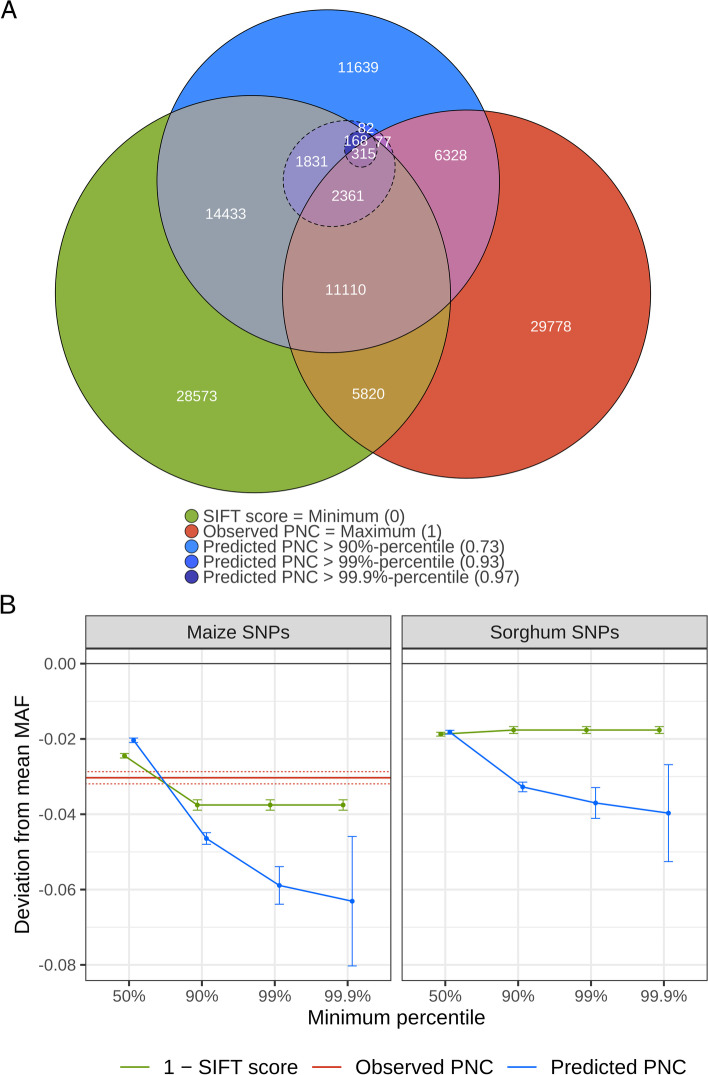
Fig. 5.
Prioritization of SNPs by SIFT score or phylogenetic nucleotide conservation (PNC). A Euler diagram of SNP sets prioritized by SIFT score = 0, observed PNC = 1, or predicted PNC > 90%, 99%, 99.9% percentile, in maize (observed polymorphisms in Hapmap 3.2.1). Concentric dashed circles refer to smaller subsets of SNPs prioritized by increasingly stringent thresholds on predicted PNC. B Decrease in minor allele frequency (MAF) of prioritized SNPs, in maize and sorghum. Difference in MAF between prioritized SNPs and all SNPs. Maize SNPs: observed polymorphisms in the Hapmap 3.2.1 reference panel [15]; Sorghum SNPs: observed polymorphisms in the reference panel of Lozano et al. [23]. SNPs were prioritized if their SIFT conservation (1 − SIFT score) or predicted PNC was above the 50%, 90%, 99%, or 99.9% percentile, or if their observed PNC was equal to 1 (tree size > 5, substitution rate < 0.05). Error bars and dotted lines represent 95% confidence intervals in two-sample t-tests, for SIFT score or predicted PNC, and observed PNC, respectively