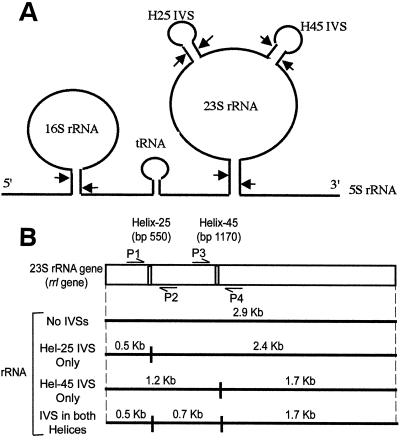
FIG. 1.
(A) Processing of the rRNA transcript, showing removal of IVSs. A typical 30S rRNA transcribed from an rrn operon is shown with 16S, spacer tRNA, 23S, and 5S rRNAs. The positions of helix 25 IVS (indicated as H25) at bp 550 and helix 45 IVS (indicated as H45) at bp 1170 are also shown as stem-loop structures on the 23S rRNA. The arrows indicate sites of cleavage by RNase III (an enzyme responsible for primary processing of the nascent 30S rRNA in E. coli and S. typhimurium [6]). (B) rrl gene for 23S rRNA, indicating the positions of helix 25 (ca. bp 550) and helix 45 (ca. bp 1170). Also indicated are primers used for PCR amplification; primers P1 and P2 amplify the helix 25 region, and primers P3 and P4 amplify the helix 45 region. The rRNA fragmentation patterns expected from excision of IVSs are also shown. The presence of 2.4- and 0.5-kb fragments indicates that an rrl gene contains an IVS in helix 25 only; 1.7- and 1.2-kb fragments indicate an IVS in helix 45 (ca. bp 1170) only, and 1.7-, 0.7-, and 0.5-kb fragments indicate IVSs in both helices. Mature 23S rRNA that lacks IVSs is 2.9 kb.