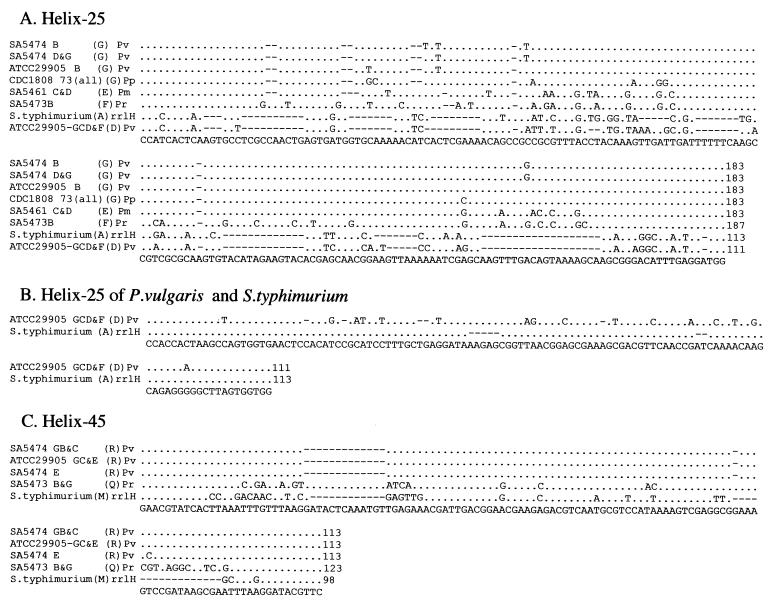
FIG. 3.
Intervening sequences from individual genes for rRNA of selected Proteus, Providencia, and Salmonella strains. Individual rrl genes were sequenced as described in Materials and Methods, and aligned using CLUSTAL X. The conserved residues are shown by dots, and deletions are shown by spaces. The consensus sequence is at the bottom. Each sequence is identified by the strain number and the I-CeuI fragment from which it was isolated, except in the case of serovar Typhimurium, where the individual rrl gene is indicated. Letters in parentheses indicate the family of IVSs to which each sequence belongs, where a family consists of sequences with 90% or greater nucleotide identity based on DNASIS (see Tables 3 and 4). We propose four families for helix 25 and two families for helix 45. The species are as follows: Pv, Proteus vulgaris; Pp, Proteus penneri; Pm, Proteus mirabilis; Pr, Providencia rettgeri, and S. typhimurium, Salmonella serovar Typhimurium. Numbers at the end of each sequence indicate the base pairs in the IVS.