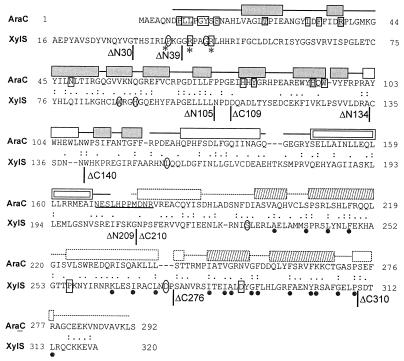
FIG. 1.
Sequence alignment of E. coli AraC (accession no. P03021) and P. putida XylS (P07859) proteins. Identical residues are marked by colons and similar ones by periods. An 18.5% identity and a 51.2% similarity were found in a 248-residue overlap (amino acids 45 to 284 in AraC and 79 to 319 in XylS). A 21.2% identity and a 58.6% similarity were found in a 104-residue C-terminal region (amino acids 183 to 284 in AraC and 218 to 319 in XylS). Sequences were aligned by using the LALIGN program. Vertical lines indicate the endpoints of XylS truncations generated in this study. Mutations of boxed XylS residues affect effector specificity. Mutations of rounded XylS residues produce semiconstitutive transcriptional activation, and mutations of residues marked by an asterisk increase XylS stability. Mutations of R41 and R45 in XylS cause both semiconstitutive and altered effector specificity phenotypes (20, 27, 37). Dots under the sequence of XylS indicate conserved residues in the AraC-XylS family (7). The AraC residues, which interact directly with arabinose, are printed on a gray background, and those interacting indirectly are boxed. The elements of the AraC NTR secondary structure are shown above the sequence. Gray boxes indicate beta-sheets, and transparent boxes indicate helices. The dimerization helix, participating in a coiled coil, is shown by a double box (32). The linker region of AraC is underlined (4, 5). The secondary structure elements of homologous MarA protein are shown above the AraC CTD sequence with a dotted outline; the helix-turn-helix motifs are shaded (29).