Fig. 1.
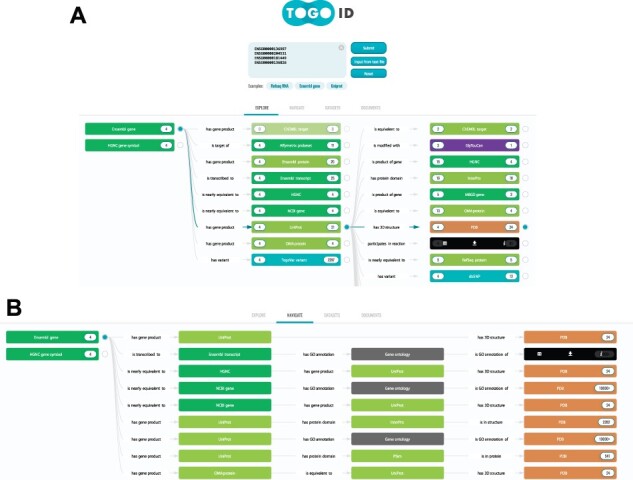
Two methods to find a proper route for converting the input IDs. (A) The screenshot of the ‘EXPLORE’ mode. By clicking the ‘Ensembl gene’ button in the examples, four gene IDs of the Yamanaka factors (Takahashi and Yamanaka, 2006) are filled in to suggest candidate datasets. By selecting a circle next to a dataset, available conversion paths will be shown. Each dataset box shows the menu by hovering the cursor. Clicking the table icon on the left opens the ‘Results’ window, where users can preview the conversion results and download in the designated output format (see Fig. 2A). The download icon in the middle is a shortcut to directly download the converted ID list. The information icon (i) on the right is to show the details of the dataset and the linked datasets. (B) In the ‘NAVIGATE’ mode, users can specify the conversion target from the pull-down menu of the supported datasets in TogoID. The candidate conversion routes will be listed
