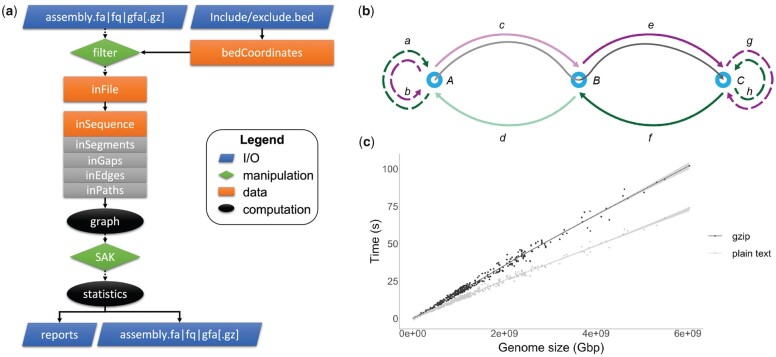
Fig. 1.
(a) Schematic of gfastats workflow. Inputs (top trapezoids) include genome assemblies in FASTA, FASTQ, GFA [.gz] formats and include/exclude lists as bed coordinate files for filtering (first diamond). These are represented internally by multiple C++ classes including their constituent elements (rectangles). The assembly can be converted to a graph (first oval), to ease manipulation by the internal Swiss Army Knife (SAK; second diamond), and then summary statistics are computed (second oval). A variety of outputs can be generated, such as summary statistics and new sequences in *fa* format. (b) Internal bidirected graph representation of the input sequences. Segments (nodes A, B, C) are connected by forward (b, c, e, g) or backward (a, d, f, h) gaps edges. Terminal nodes can optionally be associated with gaps (dashed lines, gap edges a, b, g and h). An assembly scaffold is a path in the graph (e.g. A → c → B → e → C, grey middle line). Sequence manipulation can be achieved using the internal SAK. For instance, the given path could be split by removing gap edges c and d that connect segment nodes A and B, leading to a disconnected node A, and two connected nodes B and C linked by edges e and f (portion of the path in light grey removed). Overlap edges can be treated in the same way. (c) Evaluation of gfastats runtime. Performance time is a function of genome size, with gfastats runtime increasing linearly. There is a small increase in time when handling gzip-compressed files