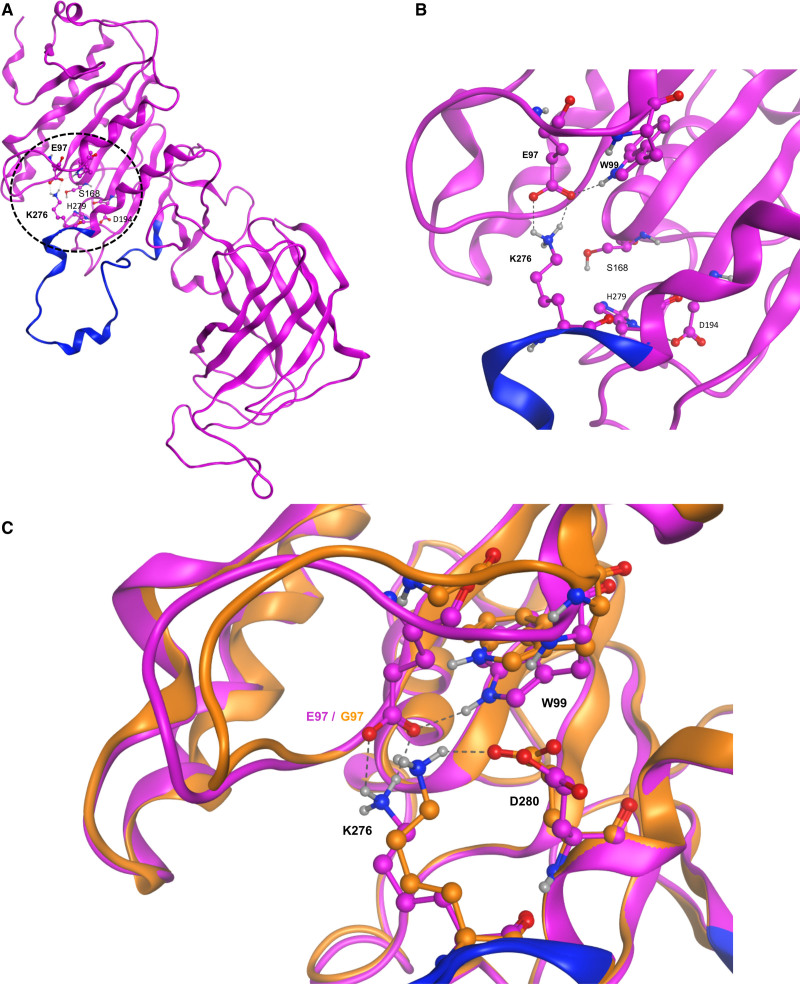
Figure 3.
E97G modifies the structural conformation of HL. A, Global structure of the homology model of hepatic lipase (HL). The N and C termini are on the top left and right, respectively. Dashed circle indicates the zone of particular interest in this study, with the location of the E97 and K276 residues and of the amino acids of the catalytic triad (S168, D194, and H279). B, Closer view of the structural motif (salt bridge and hydrogen bond) involving E97, W99, and K276. Residues of the lid domain are shown in blue. C, Detailed view of the surroundings of the K276 residue in the E97G mutant model (orange) and the HL homology model (purple) The name of some residues is indicated for clarity. Dashed lines represent the interactions (salt bridge, hydrogen bonds) between the various residues. Residues of the lid domain are shown in blue.