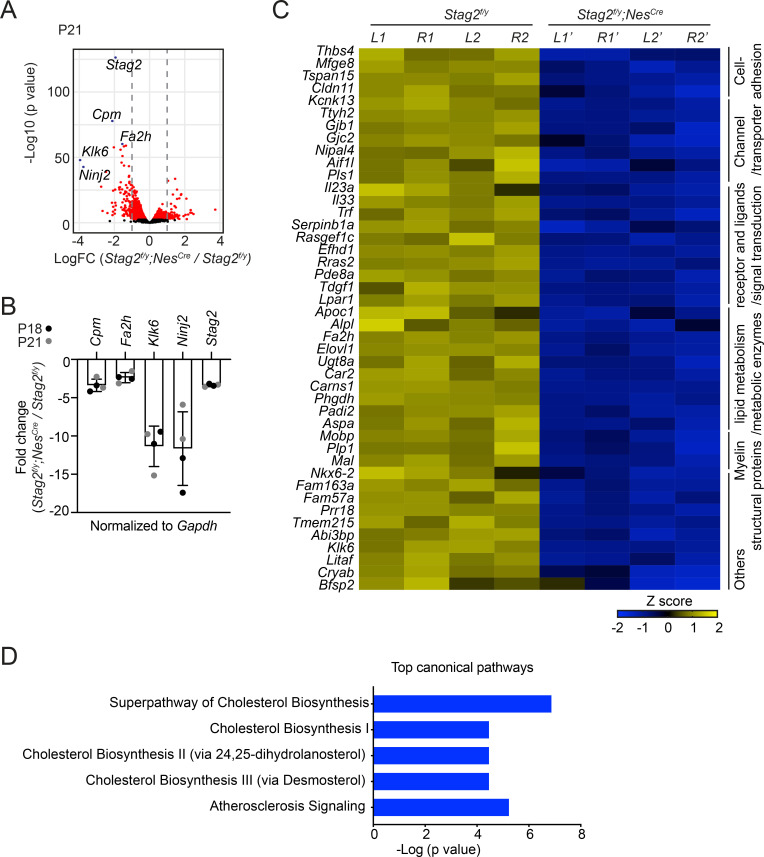
Figure 2. Stag2 ablation in mouse brains downregulates the expression of myelin genes.
(A) Volcano plot of bulk RNA-sequencing results of Stag2f/y and Stag2f/y;NesCre brain extracts. Top differentially expressed genes (DEGs) are colored blue and labeled. n = 4 pairs of P21 Stag2f/y and Stag2f/y;NesCre brain hemispheres were used for the comparison. (B) Reverse transcription quantitative PCR (RT-qPCR) analysis of the top downregulated genes in the brain extracts. n = 4 pairs of Stag2f/y and Stag2f/y;NesCre littermates were used. Mean ± standard deviation (SD). (C) Heatmap of the expression of myelin-enriched genes that were downregulated by more than twofolds in Stag2f/y;NesCre brains. L1 and R1, left and right brain hemispheres of the Stag2f/y#1 mouse. L2 and R2, left and right brain hemispheres of the Stag2f/y#2 mouse. L1’ and R1’, left and right brain hemispheres of the Stag2f/y;NesCre #1 mouse. L2’ and R2’, left and right brain hemispheres of the Stag2f/y;NesCre#2 mouse. The biological pathways of these genes are labeled on the right. (D) The top 5 canonical pathways identified by ingenuity pathway analysis (IPA) of the DEGs. The complete gene list is used as the background.