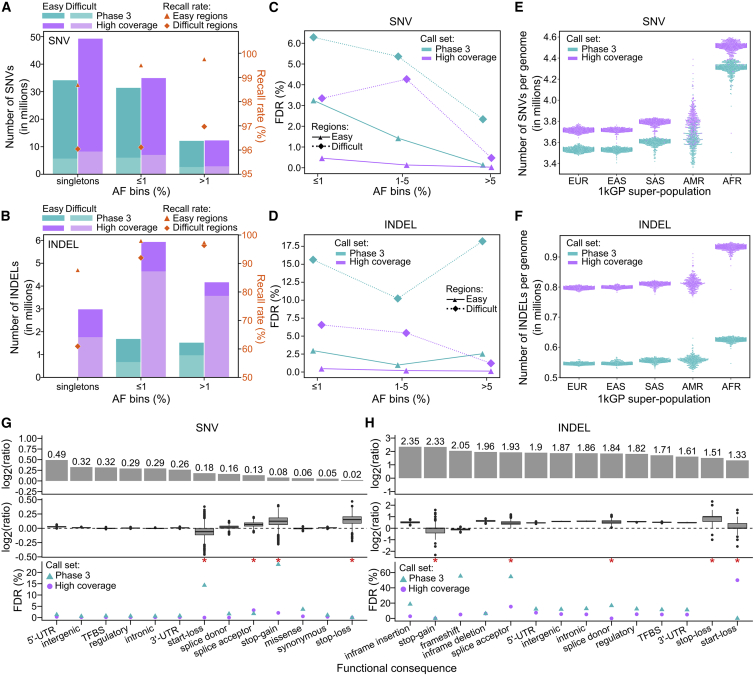
Figure 3.
Comparison of small variant calls to the phase 3 call set
(A and B) Number of SNVs (A) and INDELs (B) across the 2,504 samples in phase 3 and high-coverage datasets, stratified by AF bins and regions of the genome. Secondary y axis: % of autosomal phase 3 variants recalled in the high-coverage call set across SNVs (A) and INDELs (B) in easy and difficult regions of the genome. See also Figure S4C.
(C and D) Comparison of FDR across SNVs (C) and INDELs (D) between the high-coverage and phase 3 call sets, stratified by AF bins and regions of the genome. See also Figure S4B.
(E and F) Sample-level SNV (E) and INDEL (F) counts in the phase 3 versus high-coverage call sets, stratified by 1kGP super-population ancestry. EUR, European; AFR, African; EAS, East Asian; SAS, South Asian; AMR, American. Reported counts are at a locus level.
(G and H) Comparison of predicted functional SNV (G) and INDEL (H) counts in the high-coverage versus phase 3 call set. Log2(ratio) denotes ratio of variant counts in the high-coverage versus phase 3 call set. Top row: cohort-level comparison. Middle row: sample-level comparison. Bottom row: comparison of FDR. Red asterisks mark categories with fewer than 100 sites in sample NA12878 (i.e., categories where FDR estimation is less reliable). See also Figures S4D and S4E.
FDR in (C), (D), (G), and (H) was estimated based on comparison of calls in sample NA12878 to the GIAB truth set v3.3.2. (A), (B), and (E–H): chromosomes (chr) 1–22; (C) and (D): chr1–22 and X.