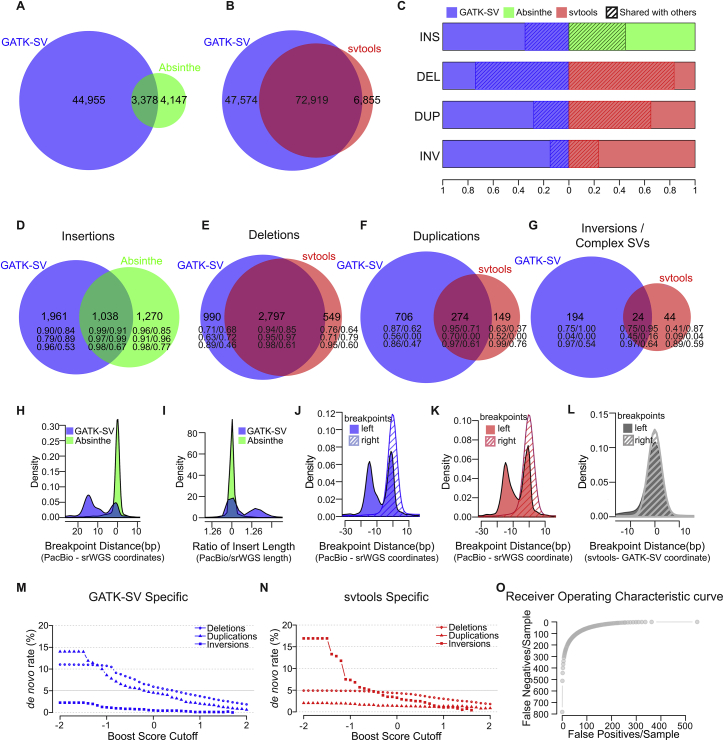
Figure S3.
Benchmark of GATK-SV, svtools, and Absinthe, related to Figure 2
(A) Overlap of insertion sites between GATK-SV and Absinthe call sets. (B) Overlap of SV other than insertions between the GATK-SV and svtools call set. (C) Overlap of SV sites of each type between GATK-SV, svtools, and Absinthe. (D) Overlap of insertions in each genome between GATK-SV and Absinthe. (E-G) Overlap of deletions (E), duplications (F), inversion and complex SVs (G) in each genome between GATK-SV and svtools. The integers in (D-G) represent count of SVs per sample, followed by proportion of SVs validated by VaPoR/proportion of SVs assessable by VaPoR in the second row, proportion of SVs supported by PacBio SVs in Ebert et al., (2021)/proportion of SVs supported by PacBio SVs in Chaisson et al. (2019) in the third row, and transmission rate/rate of biparentally inherited SVs in the fourth row. (H-I) Precision of the insertion breakpoint (H) and length (I) assessed against PacBio assemblies. (J-K) Precision of the SV breakpoints in GATK-SV (J) and svtools (K) call sets assessed against PacBio assemblies. (L) Breakpoint distance of SVs shared by GATK-SV and svtools. (M-N)de novo rate of SVs in GATK-SV (M) and svtools (N) call set when filtered at different boost score cutoffs. (O) False positives and false negatives in the GATK-SV and svtools call sets when filtered at different boost score cutoffs