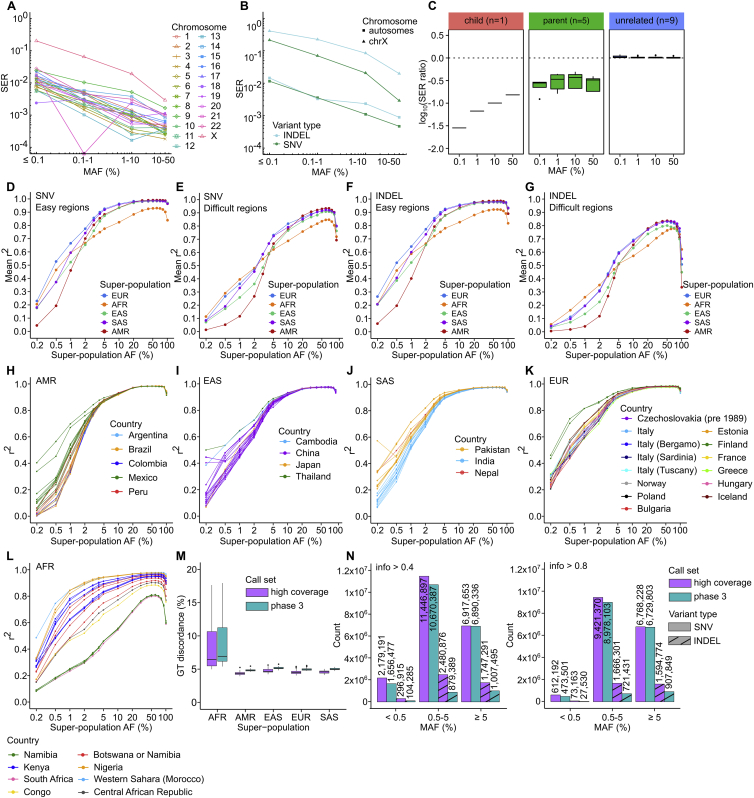
Figure S6.
SNV/INDEL phasing and imputation performance, related to Figure 5
SER: switch error rate stratified by (A) chromosome and (B) variant type. Note: SER on chr21 in the 0.1–1% MAF bin is equal to 0 (i.e. no switch errors found). This is a fluctuation due to low variant counts per MAF bin in sample NA12878 as chromosomes get smaller. Chromosome X is shown separately in (B) as it was phased using a different strategy than autosomes (statistical phasing vs. statistical phasing with pedigree-based correction, respectively). (C) Impact of inclusion of trios on the phasing accuracy of the 1kGP high-coverage call set, stratified by relationship status in the 3,202-sample cohort. log10(SER ratio) refers to the ratio of SER in the phasing run including trios (n = 3,202 samples) vs. phasing run without trios (n = 2,504 samples), computed relative to the HGSVC truth set (1 child, 5 parents, 9 unrelated samples). Imputation accuracy of the high-coverage panel stratified by super-population for SNVs (D, E) and INDELs (F, G) in easy and difficult regions of the genome. Imputation accuracy was estimated as described in Figure 5D. (H-L) Imputation accuracy of the high-coverage panel for each of the five super-populations, stratified by the population. (M) Genotype discordance rates for SNVs and INDELs imputed using the high-coverage and phase 3 panels stratified by super-population. (N) Counts of SNVs and INDELs imputed in the SGDP study dataset using the high-coverage vs. the phase 3 reference panel at info >0.4 (left) and info >0.8 (right) across three MAF bins (MAF based on the 110 imputed SGDP samples). Panels C-N are based on autosomes