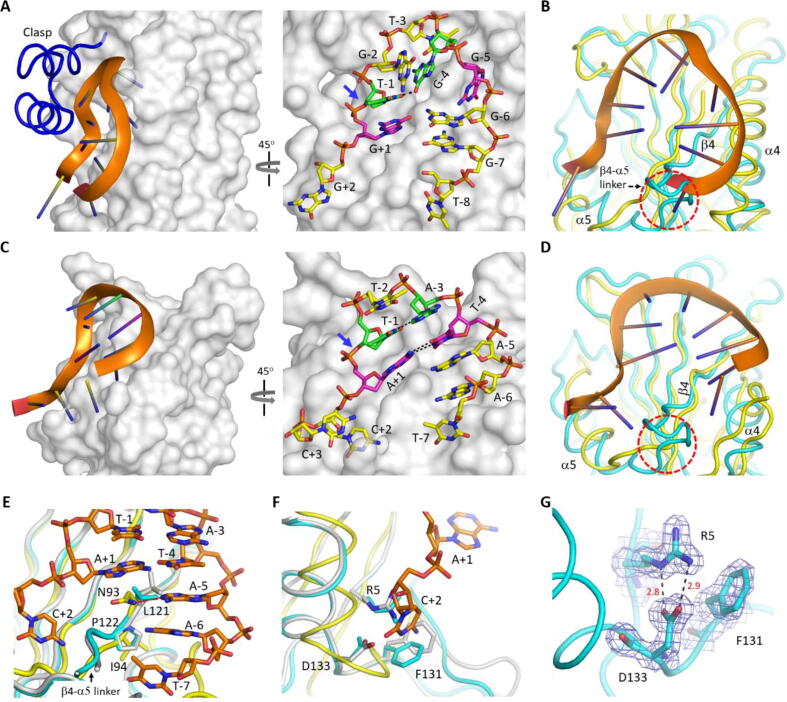
Fig. 5.
Structural comparison with other HUH-endonucleases. A) Crystal structure of TraI/ssDNA complex (PDB_ID: 2A0I). TraI protein is shown as surface with ‘clasp’ subdomain highlighted as cartoon in blue. ssDNA is shown as cartoon or sticks. B) Superposition of the core domains of NS1_Nuc and TraI relaxase. C) Crystal structure of WDV Rep/ssDNA complex (PDB_ID: 6WE1). D) Superposition of the core domains of NS1_Nuc and WDV Rep. E-F) Close view showing the clash between NS1 residues (Leu121, Pro122 and Arg5) and the nucleotides (A-5, A-6 and C + 2) of the DNA bound by WDV Rep. G) Conformation and stable interactions of Arg5 and Asp133 residues of NS1_Nuc. The 2Fo-Fc electron density maps were contour at 1.5σ level. In panels B and D-G, C-atoms are colored in cyan and white in the Form I and Form II NS1_Nuc structures, respectively. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)