Figure 3.
MFN2 interacts with STAT6 in OMM
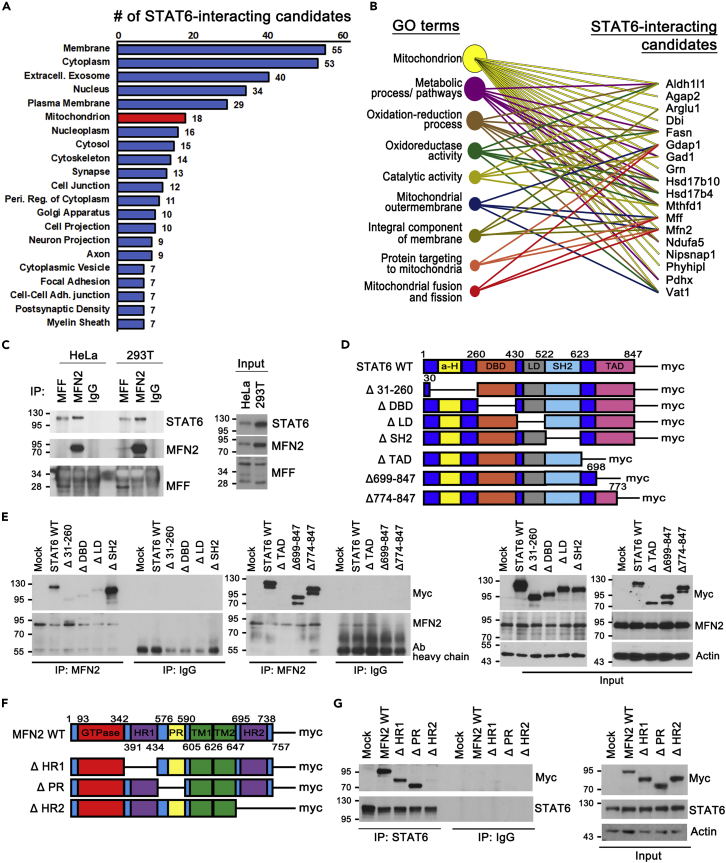
(A and B) Quantitative label-free mass spectrometry identification of STAT6-binding candidates using STAT6 IP product obtained from mouse brain.
(A) Subcellular localization of 89 STAT6-interacting candidates. Those significantly associated (p < 0.05) lists are plotted with the numbers of the binding candidates for each GO term. The STAT6-binding candidates in mitochondrion are marked with red. GO terms with less than 7 binding candidates are not plotted.
(B) The 18 STAT6-binding candidates in mitochondrion were analyzed by DAVID functional annotation corresponding to GO terms, biological function in mitochondria.
(C) Representative Western blot of immunoprecipitation using lysates obtained from HeLa and 293T.
(D) Schematic of STAT6-WT and deletion mutants with myc-tag.
(E) Western blot of immunoprecipitation using lysates from STAT6-WT or deletion mutants-expressing 293T cells.
(F) Schematic of MFN2-WT and deletion mutants with myc-tag. (HR: Heptad region, PR: Proline rich domain, TM: Transmembrane domain).
(G) Western blot of immunoprecipitation using lysates from MFN2-WT or deletion mutants-expressing 293T cells.