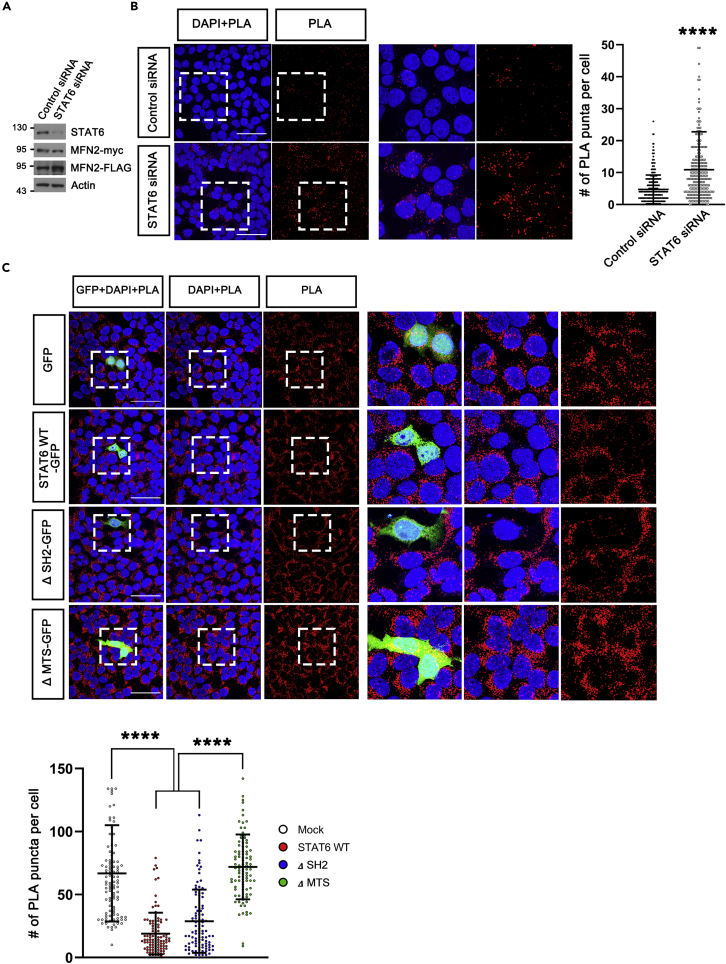
Figure 4.
STAT6 inhibits MFN2 dimerization
To analyze MFN2 dimerization, MFN2-myc/MFN2-FLAG-coexpressing stable cell line was generated.
(A) Western blot analysis using MFN2-myc/MFN2-FLAG-coexpressing stable cell line transfected with Control siRNA or STAT6 siRNA.
(B) In situ Duolink proximity ligation assay (PLA) in MFN2-myc/MFN2-FLAG-coexpressing stable cell lines transfected with Control siRNA or STAT6 siRNA. MFN2 dimerization was detected using the PLA method. PLA signals (in red) were quantified and displayed. Data are presented as means ± SD of 3 independent experiments. (Control siRNA, n = 284 and STAT6 siRNA, n = 259. ∗∗∗∗p < 0.0001), unpaired Student t-test.
(C) MFN2 dimerization was detected in MFN2-myc/MFN2-FLAG-coexpressing stable cell lines transfected with empty vector, STAT6 WT, ΔSH2, or ΔMTS (mitochondrial targeting sequences-deleted STAT6) with GFP. Data are presented as means ± SD of 3 independent experiments. (GFP, n = 105. STAT6 WT-GFP and ΔSH2-GFP, n = 100. ΔMTS-GFP, n = 101. ∗∗∗∗p < 0.0001) One-way ANOVA with Scheffe’s post-hoc test.