Figure 5.
STAT6 in OMM impairs mitochondrial fusion
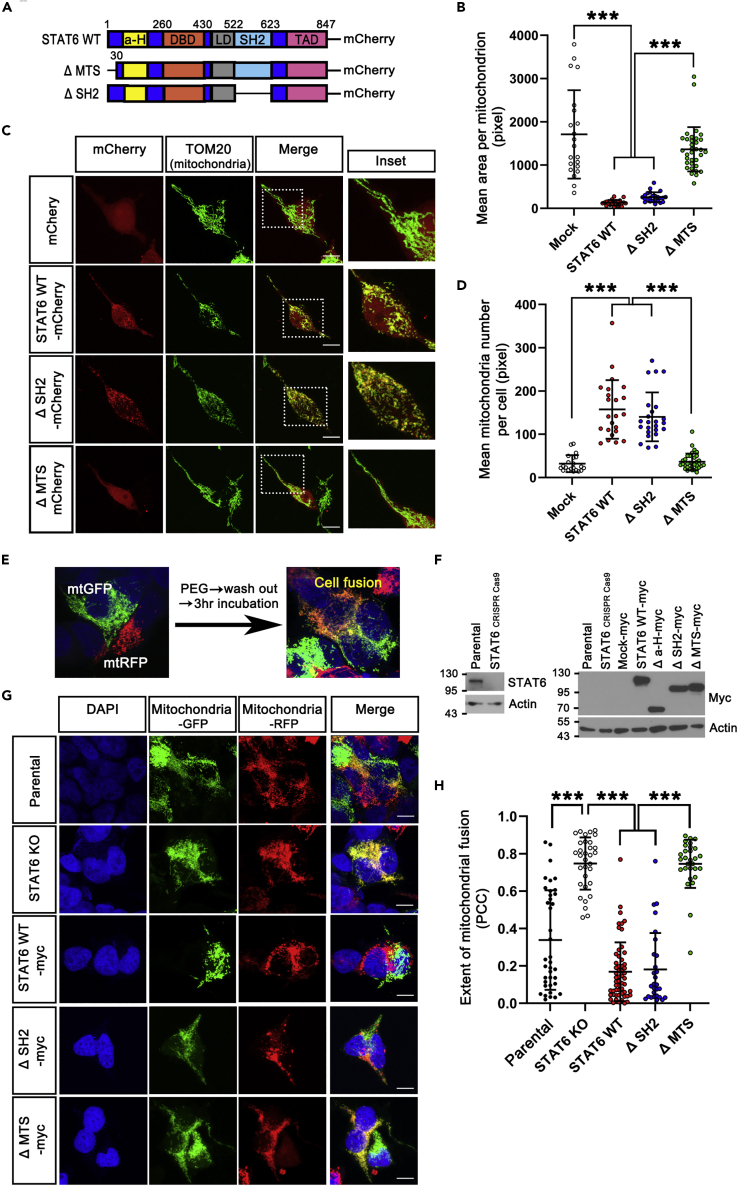
(A) Schematic of STAT6 WT and deletion mutants with mCherry.
(B–D) Confocal images (C) of mitochondrial morphology in STAT6 WT or deletion mutants-expressing 293T cells stained with TOM20. The insets show a magnification of the selected region (white square). Mitochondrial number per cell (D) and mean area per mitochondrion (B) were quantified from mCherry + cells using ImageJ. (mCherry, n = 21. STAT6 WT-mCherry, n = 23. ΔSH2-mCherry, n = 24. ΔMTS-mCherry, n = 34). Data are presented as means ± SD of 3 independent experiments. (∗∗∗p < 0.001). One-way ANOVA with Scheffe’s post-hoc test.
(E) Illustration of polyethylene glycol (PEG)- mediated cell fusion in 293T transfected with mitochondrial targeted-GFP (mtGFP) or mitochondrial targeted-RFP (mtRFP).
(F) To determine mitochondrial fusion inhibited by STAT6, STAT6-deficient cell line (STAT6 KO) was generated by CRISPR Cas9-mediated knockout from Parental cells (293T). And then STAT6 KO cell line was rescued with empty vector (Mock-myc), domain-deleted STAT6 mutants, and wild-type STAT6 with myc-tag (STAT6 WT-myc). Cell extracts obtained from the stable cell lines mentioned above were analyzed by Western blot using indicated antibodies.
(G) Confocal images of mitochondrial fusion in HEK 293T polykaryons subjected to the PEG-based fusion assay in the stable cell lines mentioned above. These stable cells were subsequently transfected with mtGFP or mtRFP and then co-cultured, treated with PEG for cell fusion. Mitochondrial fusion is indicated by colocalization (yellow mitochondria) of mitoGFP and mitoRFP.
(H) Quantitative analysis of the extent of mitochondrial fusion in individual hybrid cells was performed by the Pearson’s correlation coefficient (PCC). (Parental, n = 39. STAT6 KO, n = 33. STAT6 WT-myc, n = 57. ΔSH2-myc, n = 29. ΔMTS-myc, n = 30). Data are presented as means ± SD of 3 independent experiments. (∗∗∗p < 0.001). One-way ANOVA with Scheffe’s post-hoc test.