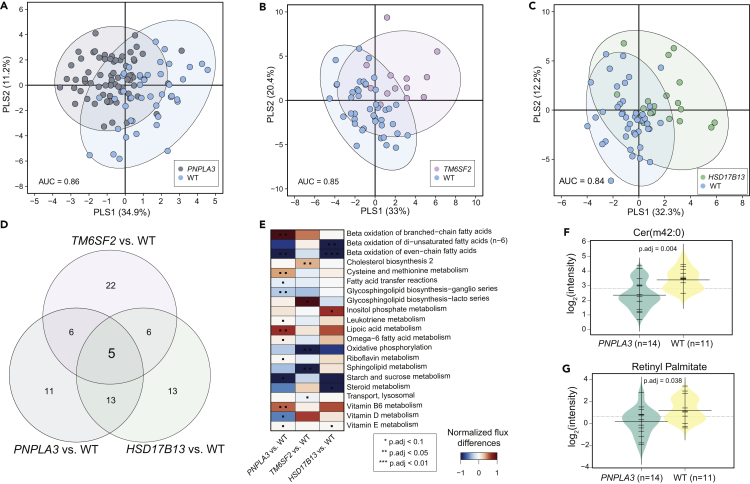
Figure 6.
Metabolic pathway differences across the genetic modifiers of NAFLD
PLS-DA–score plot showing metabolic pathway differences across three major gene variants vs. WT groups.
(A) PNPLA3 vs. WT.
(B) TM6SF2 vs. WT.
(C) HSD17B13 vs. WT.
(D) Metabolic pathways/subsystems commonly regulated among three different gene variants with (PLS-DA, VIP score >1).
(E) Normalized flux differences across the metabolic subsystems between three major gene variants associated vs. WT groups. Red and blue color denotes up- and downregulation (two-sample t-test, p < 0.05 adjusted for FDR) of reaction fluxes across the metabolic subsystems within at two.
(F and G) Beanplots showing differences in the log2 intensities of the Cer(m42:0) and retinyl palmitate measured in the serum samples of a patient subgroup (n = 14) exclusively carrying variants for PNPLA3 (homo- and heterozygous) gene vs. WT (n = 11). The dotted line denotes the mean of the population, and the black dashed lines in the bean plots represent the group mean. Statistical significance was determined by (two-sample t-test, p < 0.05 adjusted for FDR, p.adj).