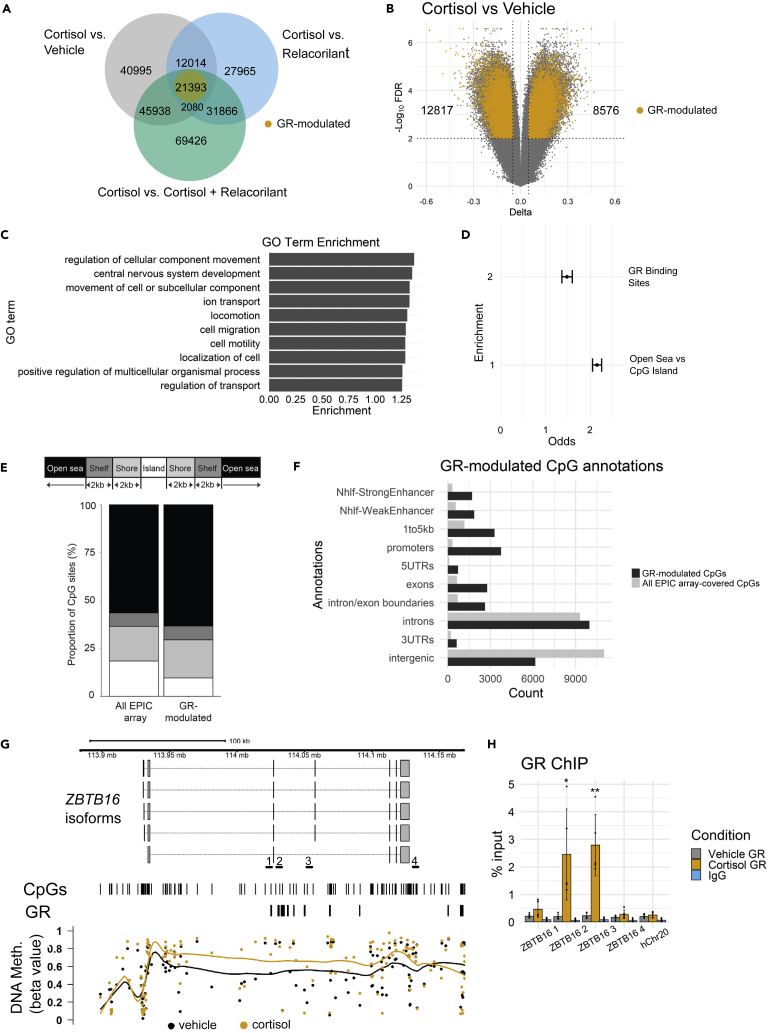
Figure 4.
Chronic stress-driven GR activation induces widespread, but genomic context-dependent, methylomic changes
(A) Venn diagram showing overlap of differentially methylated CpGs across cortisol vs vehicle, cortisol vs. relacorilant, and cortisol vs. cortisol + relacorilant comparisons. Yellow circle represents GR-modulated CpGs that show a consistent direction in methylation change across all comparisons.
(B) Volcano plot showing differences in CpG methylation levels in the cortisol vs. vehicle condition. GR-modulated CpG sites are labeled in yellow. Dotted vertical lines represent a 5% change in methylation levels. Dotted horizontal lines represent FDR < 0.01.
(C) Gene ontology (GO) biological process pathway analysis of GR-modulated genes identified in the EPIC array analysis. GO analysis was performed using WebGestalt (WEB-based Gene SeT AnaLysis Toolkit) (Liao et al., 2019). The top 10 most enriched pathways are shown.
(D) Enrichment analysis of differentially methylated CpGs localized at previously identified GR binding sites and open sea vs. CpG island sites.
(E) Proportion of CpGs for all EPIC array-covered and GR-modulated sites.
(F) Genic and enhancer annotations of GR-modulated CpG sites. GR-modulated enrichment is shown in black and all EPIC array-covered CpGs are shown in gray.
(G) Metagene plot showing locally estimated scatterplot smoothing of CpG DNA methylation values across the ZBTB16 gene. Locations of CpGs and previously identified GR binding sites are indicated. Yellow points represent DNA methylation in cortisol-treated IMR-90 cells. Black points represent DNA methylation in vehicle-treated IMR-90 cells. Primer locations used for ChIP-qPCR are indicated.
(H) ChIP-qPCR analysis across the ZBTB16 gene. Bars represent the average of 5 biological replicates. Error bars represent one SD above and below the mean. ∗p < 0.05; ∗∗p < 0.01; ∗∗∗p < 0.001; ∗∗∗∗p < 0.0001.