Abstract
The global regulator LaeA controls secondary metabolism in diverse Aspergillus species. Here we explored its role in regulation of itaconic acid production in Aspergillus pseudoterreus. To understand its role in regulating metabolism, we deleted and overexpressed laeA, and assessed the transcriptome, proteome, and secreted metabolome prior to and during initiation of phosphate limitation induced itaconic acid production. We found that secondary metabolite clusters, including the itaconic acid biosynthetic gene cluster, are regulated by laeA and that laeA is required for high yield production of itaconic acid. Overexpression of LaeA improves itaconic acid yield at the expense of biomass by increasing the expression of key biosynthetic pathway enzymes and attenuating the expression of genes involved in phosphate acquisition and scavenging. Increased yield was observed in optimized conditions as well as conditions containing excess nutrients that may be present in inexpensive sugar containing feedstocks such as excess phosphate or complex nutrient sources. This suggests that global regulators of metabolism may be useful targets for engineering metabolic flux that is robust to environmental heterogeneity.
Keywords: Aspergillus pseudoterreus, Itaconic acid, laeA, Process robustness, Multi-omics, Phosphate
Highlights
-
•
The Itaconic acid biosynthetic gene cluster is regulated by laeA.
-
•
LaeA is required for production of itaconic acid.
-
•
Overexpression of laeA attenuates genes involved in phosphate acquisition.
-
•
Global regulator engineering increases robustness of itaconic acid production.
1. Introduction
Itaconic acid is a five-carbon dicarboxylic acid that has been recognized as a platform chemical with broad applications in the production of commodity and specialty chemicals such as polymers, coatings and solvents (W et al., 2004). Itaconic acid is produced naturally from sugars by the filamentous fungus Aspergillus terreus during submerged fermentation culture at high titer and low pH (Larsen and Eimhjellen, 1955; Eimhjellen and Larsen, 1955). Significant market opportunities are available for the development of itaconic acid as a biorenewable product, however to be competitive with petrochemical-derived products it was estimated in 2004 that the fermentation cost needed to be at or below $0.25/pound (W et al., 2004). The primary pathways to achieve this were identified as increased fermentation rate, improvement of final titer, and production from inexpensive C5 sugars from lignocellulosic feedstocks. Since then, demand has remained low.
Production of itaconic acid by Aspergillus terreus on glucose occurs via glycolysis and the tri-carboxylic acid cycle (Willke and Vorlop, 2001). Cis-aconitate is released by aconitase during isomerization of citrate to isocitrate and transported out of the mitochondrion by the mitochondrial carrier protein mttA (Steiger et al., 2016). Cis-aconitate is then decarboxylated by the cis-aconitate decarboxylase (cadA) to itaconate in the cytosol. Deletion of the major facilitator superfamily protein from the itaconic acid biosynthetic cluster (mfsA) reduces, but does not eliminate, itaconic acid production (Deng et al., 2020) while overexpression improves productivity (Li et al., 2011) suggesting it plays a role as the major, but not sole, transporter for itaconate across the plasma membrane (Shin et al., 2017). The genes encoding this metabolic pathway are colocalized on the chromosome suggesting their regulation may be coordinated as is the case for many secondary metabolite pathways in fungi (Li et al., 2011).
Overall titer, yield, and production rate have been improved by systematic alteration of culture conditions for high productivity from glucose (Hevekerl et al., 2014a, 2014b; Karaffa et al., 2015; Krull et al., 2017; Kuenz and Krull, 2018) while utilization of pentose sugars as well as the impact on productivity of nutrients and growth inhibitors present in lignocellulosic feedstocks (Saha and Kennedy, 2017, 2018, 2020; Saha et al., 2017, 2019; Kollath et al., 2019; Sandor et al., 2021) has been investigated. Limited efforts to produce itaconic acid from lignocellulosic feedstocks such as enzymatically saccharified wheat straw hydrolysate have generally yielded poor results due the sensitivity of the process to impurities in the culture medium such as acetic acid and furfural (Kuenz and Krull, 2018; Saha et al., 2019) and the need to maintain phosphate (Willke and Vorlop, 2001) and manganese (Karaffa et al., 2015; Saha et al., 2019) limited conditions for high productivity. High yields of itaconic acid production have been achieved using fungi (Xie et al., 2020; Sun et al., 2020; Becker et al., 2020; Zhao et al., 2018, 2019; Hosseinpour Tehrani et al., 2019) which are capable of producing itaconic acid from purified hexose and pentose sugars characteristic of the sugars present in lignocellulosic feedstocks at pH well below the pKa of itaconic acid (pKa = 3.58) allowing for production of the free acid (Bafana and Pandey, 2018). However, strategies are needed to improve the robustness of itaconic acid production to compositional variance present in lignocellulosic feedstocks since utilization of pure sugars, purified molasses, or starch hydrolysates are not economically competitive when compared to raw materials from the petrochemical industry (Kuenz and Krull, 2018).
Bioconversion processes often fail when moved to non-ideal conditions in part because the organism responds to the new environment by altering the expression of genes which subsequently changes flux through the organism's metabolic network. We hypothesized that altering the organism's ability to respond to environmental perturbation, by modification of global regulatory mechanisms, would improve robustness of the bioprocess to environmental change. In many fungi the putative methyltransferase laeA is a global regulator of genes present in secondary metabolite clusters and the response to environmental ques (Bayram and Braus, 2012; Amare and Keller, 2014; Gerke and Braus, 2014). In Trichoderma reesei, laeA regulates expression of cellulases and polysaccharide hydrolases (Karimi Aghcheh et al., 2013a, Karimi-Aghcheh et al., 2013b) as well as a variety of secondary metabolite clusters (Karimi Aghcheh et al., 2013a, Karimi-Aghcheh et al., 2013b), while in Aspergillus nidulans, LaeA forms a complex with VeA and VelB that affects the production of secondary metabolites and participates in light-responsive developmental regulation (Bayram et al., 2008). LaeA has also been implicated in regulation of central carbon metabolism and citric acid production in Aspergillus carbonarius (Linde et al., 2016) Aspergillus niger (Niu et al., 2015), and Aspergillus luchuensis (Kadooka et al., 2020). Here we examined the impact of laeA on production of itaconic acid, characterized its impact as a global regulator of gene expression, and examined its utility for bioconversion of lignocellulosic feedstocks by Aspergillus pseudoterreus.
2. Methods
2.1. Cultivation conditions
A. pseudoterreus strain ATCC®32359™ was obtained from American Type Culture Collection (Manassas, Virginia). Overexpression and deletion strains of laeA were described previously (Dai and Baker, 2016). All strains were maintained on potato dextrose agar (BD, USA). Spore inoculum was grown on potato dextrose agar at 30C and harvested after 5 days. Transformants were selected on minimum medium agar (10 g/L glucose, 6 g/L Na2NO3, 0.52 g/L KCL, 0.52 g/L MgSO4.7H2O, 1.7 g/L KH2PO4, 0.1 mg/L Biotin, 0.1 mg/L pyridoxine-HCl, 0.1 mg/L thiamine-HCl, 0.1 mg/L riboflavin, 0.1 mg/L para-amino benzoic acid, 0.1 mg/L nicotinic acid, 22 mg/L ZnSO4.7H2O, 11 mg/L H3BO3, 5 mg/L MnCl2.4H2O, 5 mg/L FeSO4.7H2O, 1.7 mg/L CoCl2.6H2O, 1.6 mg/L CuSO4.5H2O, 1.5 mg/L Na2MoO4.2H2O, 50 mg/L Na2EDTA, and 15 g/L agar) supplemented with 100 μg/ml hygromycin or 250 μg/ml bleomycin as appropriate. For pyrithiamine marker selection, the thiamine-HCl was eliminated from MM media. For shake-flask experiments, A. pseudoterreus strains were cultivated in minimal glucose/xylose medium (GX) (32.4 g/L glucose, 17.6 g/L xylose, 2.36 g/L (NH4)2SO4, 0.11 g/L KH2PO4, 2.08 g/L MgSO4.7H2O, 0.13 g/L CaCl2.2H2O, 74 mg/L NaCl, 1.3 mg/L ZnSO4.7H2O, 0.7 mg/L MnCL2.4H2O, 5.5 mg/L FeSO4.7H2O, 0.2 mg/L CuSO4.5H2O, pH 3.4) (Riscaldati et al., 2000) at 30°C and 200 rpm unless otherwise noted. To the GX medium 0.14 g/L KH2PO4 was added to make GX-P and GX-NP and 2 g/L yeast extract was added to make GX-N and GX-NP. Spore suspensions are maintained in 15% glycerol at −80°C. For multi-omic analyses spores from wild-type (ATCC®32,359™) and laeA overexpressing A. pseudoterreus were inoculated at 106/mL into 50 mL production medium (PM) (100 g/L glucose, 2.36 g/L (NH4)2SO4, 0.11 g/L KH2PO4, 2.08 g/L MgSO4.7H2O, 0.13 g/L CaCl2.2H2O, 74 mg/L NaCl, 1.3 mg/L ZnSO4.7H2O, 0.7 mg/L MnCL2.4H2O, 5.5 mg/L FeSO4.7H2O, 0.2 mg/L CuSO4.5H2O, pH 3.4) (Riscaldati et al., 2000) in 250 mL smooth-walled Erlenmeyer flasks at 30C and 200 rpm in an orbital shaker.
2.2. Strain construction
Over-expression and deletion cassettes for A. pseudoterreus laeA were described previously (Dai and Baker, 2016). Briefly, the double-joint PCR method (Yu et al., 2004) was applied to prepare the laeA deletion construct with oligos P1 to P8. Oligo pair P1/P3 was used for PCR isolation of the upstream region (−1177bp to −75 bp) fragment of the laeA gene, P4/P5 for the hph marker gene, P6/P8 for the downstream region (955 bp to 2255 bp) fragment of laeA gene, and P2/P7 for the entire laeA deletion cassette from the double-joint PCR fragment. The laeA over-expression construct was prepared with oligos 15 to 24 using Gibson assembly (Gibson et al., 2009). The oligo pairs P15/P16 and P17/P18 were used to isolate the gpdA (M33539.1) promoter and the laeA (AY394722.1) coding region of Aspergillus nidulans, which were fused together by PCR. The 5′-upstream fragment of the A. niger pyrG gene (P19/P20), the gpdAp-laeA fragment (P21/P22), A. nidulans TrpC transcriptional terminator (P23/P24), Aspergillus oryzae pyrithiamine resistance (ptrA) marker gene (P25/P26), and the 3′-downstream fragment of A. niger pyrG gene (P27/P28) were prepared by PCR and fused together by yeast gap-repair cloning method (Dai et al., 2013) to form the transgene expression construct. Finally, the laeA over-expression construct for A. pseudoterreus was prepared by the yeast gap-repair cloning method. The laeA over-expression cassette was targeted to the 5′- upstream region of laeA (between −1.7 kb and −1.57 kb). Primers P29/P30 were used to amplify the laeA upstream fragment between −2716 bp and −1698bp of A. pseudoterreus, P31/P32 for the entire fragment of gpdAp-laeA-trpCt-ptrA, and P33/P34 for the laeA upstream fragment between −1571 bp and −493 bp of A. pseudoterreus prior to assembly and transformation. Deletion of laeA in transgenic strains of A. pseudoterreus was confirmed by PCR with oligo pair P1/P107 and P108/P8. Integration of the laeA over-expression cassette was confirmed by PCR with oligo pairs P23/P32 and P31/P32. Oligos are presented in Table 1.
Table 1.
Oligos used for strain construction.
| Oligo | Name | Sequence |
|---|---|---|
| P1 | LeAF1 | ACAGGTACTTCCATCTTGTACTGGT |
| P2 | LeAF2 | TCtcctccaacgtccgatct |
| P3 | LeAR3 | acctccactagctccagcaagccgaacagaggtaaagacga |
| P4 | hphF4 | tcgtctttacctctgttcggcttgctggagctagtggaggtca |
| P5 | hphR5 | taccaacgtgcgaccatttTCTcggtcggcatctactctattcct |
| P6 | LeAF6 | aggaatagagtagatgccgaccgagaaaatggtcgcacgttggta |
| P7 | LeAR7 | AAGCGTCTCTTTCCTGGGTCTT |
| P8 | LeAR8 | TGCCAGTTCTGTTGGACATCTCT |
| P15 | gpdAF | cgcagatctcaagctgtaaggatttcggca |
| P16 | gpdAR | CACCGGGCCCATCTCAAACATTGTGATGTCTGCTCAAGCG |
| P17 | laeAF | cgcttgagcagacatcacaatgtttgagatgggcccggtg |
| P18 | laeAR | cgcagatctGAGGATTATGAGAAGGGAGC |
| P19 | pyrG5F | GTAACGCCAGGGTTTTCCCAGTCACGACGtttaaacATGCATCATTCTCCCGCTTTGT |
| P20 | pyrG3R | tgccgaaatccttacagcttgAAGCTTcatcgccaatcacctcaatcac |
| P21 | LaeA5F | gtgattgaggtgattggcgatgAAGCTTcaagctgtaaggatttcggca |
| P22 | LaeA3R | acttctacacagccatcggtccAAGCTTgaggattatgagaagggagct |
| P23 | trc5F | agctcccttctcataatcctcAAGCTTggaccgatggctgtgtagaagt |
| P24 | trp3R | cgtaatcaattgcccgtctgtcagagagcggattcctcagtctcgt |
| P25 | PTR5F | acgagactgaggaatccgctctctgacagacgggcaattgattacg |
| P26 | PTR3R | acagcagtgcttatctgcgatgacgagccgctcttgcatctttgt |
| P27 | pyrG5F | acaaagatgcaagagcggctcgtcatcgcagataagcactgctgt |
| P28 | pyrG3R | GCGGATAACAATTTCACACAGGAAACAGCgtttaaactgtgccagtcaattgtccgaagt |
| P29 | upstF | cgaggtcgacggtatcgataGTTTAAACCTCCCAGGTACCGACTAAC |
| P30 | upstR | ctcaatcacaGATCATGTTTGGGTGGGTTC |
| P31 | ElaeAF | aaacatgatcTGTGATTGAGGTGATTGGCG |
| P32 | ElaeAR | ctctgtgcctACAGCAGTGCTTATCTGCGATG |
| P33 | downT | gcactgctgtAGGCACAGAGTAACAGGTAGGTAGACAG |
| P34 | downR | agtggatcccccgggctgcaGTTTAAACTCCCACGCACGAAAGCAACT |
| P107 | hygR | GTACTTCTACACAGCCATCGGTCCA |
| P108 | hygL | CGTTATGTTTATCGGCACTTTGCAT |
2.3. Reference genome sequencing, assembly, and annotation
Genomic DNA and RNA was isolated from A. pseudoterreus (ATCC®32359™) using a yeast genomic DNA purification kit (AMRESCO, Solon, OH) and Maxwell 16 LEV Plant RNA kit (Promega, Madison, WI) respectively. Genomic DNA was sequenced by paired-end 250 base pair sequencing on an Illumina MiSeq platform (San Diego, CA) and assembled into contigs with the CLC Genomics Workbench (Qiagen, Hilden, Germany). Stranded RNA from A. pseudoterreus (ATCC®32359™) grown in YPD (10 g/L peptone,10 g/L yeast extract, 20 g/L glucose), YES (150 g/L sucrose, 20 g/L yeast extract, 50 mg/L MgSO4.7H2O, 10 mg/L ZnSO4.7H2O, 5 mg/L CuSO4.5H2O), MM, and MM-W (20 g/L wheat instead of 20 g/L glucose) medium was sequenced by paired-end 100 base pair sequencing on an Illumina HiSeq2500 platform. RNA sequences were used to produce a high-quality genome annotation with the JGI genome annotation pipeline. The assembled genome has been deposited at Genbank (Accession: PRJNA420104) and the annotated version has been made available through the MycoCosm portal (https://mycocosm.jgi.doe.gov/Asppseute1/Asppseute1.home.html) (Grigoriev et al., 2014). Orthology of genes with other Aspergillus species that have high quality annotated reference genomes was determined using SPOCS (Curtis et al., 2013). Secondary metabolite clusters were predicted using fungiSMASH 4.2.0 (Blin et al., 2017).
2.4. Multi-omic analyses
Tissue samples were flash frozen in liquid nitrogen after collection. RNA was isolated using a Maxwell 16 LEV Plant RNA kit and was sequenced by stranded single-end 50 base pair sequencing on an Illumina HiSeq2500 platform. Sequencing reads were mapped to the A. pseudoterreus coding sequence models, using Bowtie2 v2.2.8 (Langmead and Salzberg, 2012). The alignments were sorted, converted to bam format, and quantified with Samtools v1.3.1 (Li, 2011). For global extracellular metabolomics analysis, supernatants were dried, chemically derivatized, acquired and analyzed as previously reported (Pomraning et al., 2021). Specific metabolites were also quantified using a GC-MS with external calibration curves corresponding to authentic chemical standards as described previously (Pomraning et al., 2021). Absolute quantification of some metabolites was performed by HPLC. Samples were filtered with a 0.2 μm syringe filter and analyzed for 45 min using an Aminex HPX-87H ion exclusion column with a 1 mM H2SO4 flow of 0.6 ml/ml. The temperature of the column was 60C. The refractive index at 45C and the UV absorption at 210 nm were measured. Targeted and global proteomics was performed as previously described (Pomraning et al., 2021).
2.5. Data analysis
For protein samples, sample level quality was ensured by a robust Principal Component Analysis to compute a robust Mahalanobis distance based on sample-level parameters (Matzke et al., 2011). The default for normalization is standard global median centering to account for total abundance differences between samples. A test was performed to assure that these factors are not biased (Webb-Robertson et al., 2010). For this dataset there was no bias detected and we utilized global median centering (Callister et al., 2006). Protein quantification was performed with standard reference-based median averages (Polpitiya et al., 2008; Matzke et al., 2013). Statistics were performed with established standard methods (Webb-Robertson et al., 2017). For RNA samples, raw read alignment counts were assessed statistically in R v3.5 using the DESeq2 method as implemented in Bioconductor v3.8 (Love et al., 2014). For visualization purposes the aligned reads were examined in the Integrative Genomics Viewer (Robinson et al., 2011). Enrichment of gene ontology terms used FunRich (Pathan et al., 2015).
3. Results
3.1. Genome assembly and annotation of Aspergillus pseudoterreus
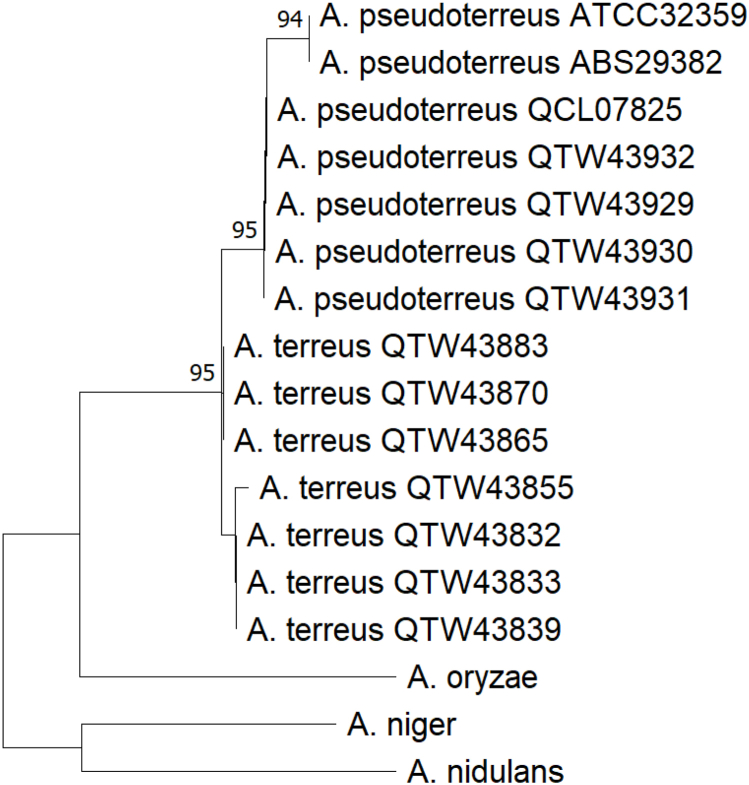
We sequenced the genome of A. terreus strain ATCC®32359™ (derived from NRRL 1960 (Nakagawa et al., 1975)) as a resource for development as a bioproduction platform organism to an average depth of 109x and assembled into 272 contigs. The resulting assembled genome sequence is 29.5 Mb (N50, 678,262 bp; Nmax, 1,693,594 bp) with G + C content of 52.3%. Stranded RNA isolated from four growth conditions (YPD, YES, MM, and MM-W) was sequenced and used to annotate the genome with the JGI annotation pipeline. Comparison of randomly selected 1 kilobase sections of the genome with other Aspergillus species by Blastn identified the recently described A. pseudoterreus as the most similar species to strain ATCC®32359™ (Samson et al., 2011). Phylogenetic analysis using the β-tubulin genes with those from other Aspergillus species confirmed this (Fig. 1). We therefore propose designation of strain ATCC®32359™ as A. pseudoterreus and have named it as such in the genome sequence assembly deposited at Genbank (Accession: PRJNA420104) and the annotated sequence available through the JGI's MycoCosm portal (https://mycocosm.jgi.doe.gov/Asppseute1/Asppseute1.home.html).
Fig. 1.
Phylogenetic analysis of A. pseudoterreus strain ATCC32359. Calmodulin coding regions from selected Aspergillus species were downloaded from ENA and NCBI and aligned using Muscle. The consensus evolutionary history was inferred from 310 positions using the Maximum Likelihood method with 500 bootstrap replicates in MEGA (Tamura et al., 2021).
3.2. LaeA regulates itaconic acid production
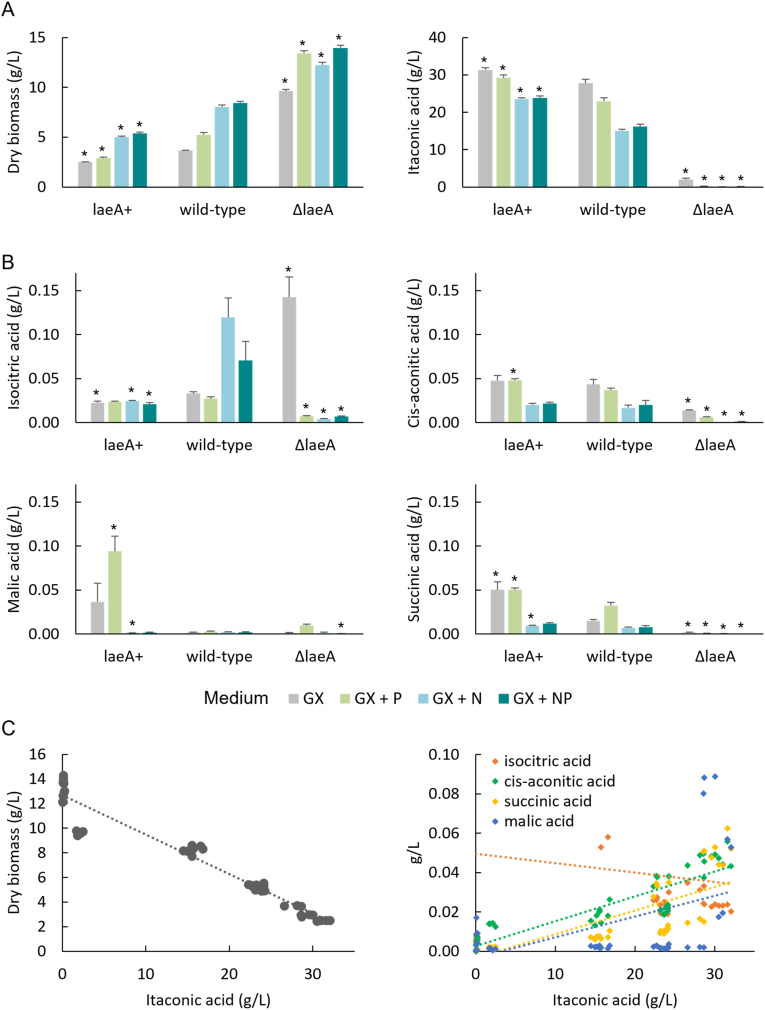
The global regulator LaeA controls secondary metabolism in a wide variety of fungi (Bayram and Braus, 2012; Amare and Keller, 2014; Gerke and Braus, 2014). To assess the impact of LaeA on metabolism in A. pseudoterreus we replaced the endogenous laeA gene with hph by targeted homologous recombination to create a deletion strain and overexpressed the laeA homolog from A. nidulans to create a laeA overexpression strain. Overexpression, deletion, and the parental wild-type strain were cultivated for eight days in shake flasks in minimal medium with glucose and xylose present at a ratio typical of a lignocellulosic feedstock (GX) to examine the effect of laeA on itaconic acid production (Fig. 2a). Deletion of laeA nearly eliminated itaconic acid production (decrease of 94%) while overexpression increased itaconic acid production by 13% on GX medium. This suggests that LaeA is important for regulating itaconic acid production. In these conditions itaconic acid is the major secreted product from the wild-type strain with less than 1 g/L detected in the culture supernatants for other acids (Fig. 2b).
Fig. 2.
Regulation of itaconic acid production by LaeA in Aspergillus pseudoterreus. Strains were cultivated in shake flasks for 8 days until all of the glucose and xylose present in the medium had been consumed from an initial concentration of 50 g/L total sugar. A) The major products of the cultivation are biomass and itaconic acid. Both are affected by laeA deletion and overexpression. B) Production of other tricarboxylic acid cycle derived organic acids at minor levels. Citric acid was not detected at > 5 mg/L in any sample. C) Correlation between itaconic acid production and final biomass or other acids. GX (glucose/xylose medium), P (phosphate), N (yeast extract). Asterisks indicate significant differences between either the laeA + or ΔlaeA strain and the wild-type in each condition.
We spiked the GX medium with phosphate, yeast extract, or both to mimic the presence of phosphate and uncharacterized nutrient sources that may be present in lignocellulosic feedstocks. Supplemental yeast extract in particular reduced itaconic acid production by 46% in the wild-type strain while additional phosphate reduced itaconic acid production by 17%. Overexpression of LaeA made bioconversion to itaconic acid more robust to these nutrient additions with only 25% and 6% decreases in yield respectively (Fig. 2a). We hypothesized that other organic acids would be produced by the laeA deletion strain in the absence of itaconic acid production, and to a small extent this was true as isocitric acid was produced at a higher level in the laeA deletion strain, however the increase was small (0.1 g/L). By far, the largest positive impact on productivity in the laeA deletion strain was in biomass production (Fig. 2a). Itaconic acid production is strongly negatively correlated with biomass production across the different mutant and medium conditions examined (R2 = 0.94) and is positively correlated with cis-aconitic acid production (R2 = 0.81) and to a lesser extent succinic acid production (R2 = 0.61) (Fig. 2c). Overall, this suggests that the processes regulated by LaeA in A. pseudoterreus promote flux away from growth and toward production of itaconic acid.
3.3. Transcriptome analysis of LaeA overexpression
We quantified transcripts by RNA-sequencing of wild-type and the LaeA overexpression strains of A. pseudoterreus during growth phase at 36 hours in shake flasks to assess global effects on gene expression. Global profiling quantified 10,228 transcripts of which 1089 are significantly up-regulated and 458 significantly down-regulated (q < 0.05, fold-change > 2x). The transcript level of laeA in the overexpression strain was 17.6x higher than in the wild-type strain. Gene ontology analysis found that in general, genes involved in metabolism are up-regulated in the LaeA overexpression strain while genes involved in translation are down-regulated (Table 2). To investigate the metabolic genes in more detail we examined those with annotations in an Aspergillus terreus metabolic model (Liu et al., 2013). Changes in expression were mapped to 211 metabolic pathways to identify those enriched for up- or down-regulated genes. Activated pathways tend to be involved in utilization of alternative carbon sources (Table 3) while the most significantly up-regulated individual metabolic genes are involved in terreic acid biosynthesis (513675, 172x; 283888, 27x) and itaconic acid biosynthesis (531880, 92x; 505023, 50x; 497765, 33x; 466479, 15x).
Table 2.
Gene ontology term enrichment in LaeA overexpression strain transcriptome. Significantly up- and down-regulated genes in the LaeA overexpression strain versus wild-type during growth phase were assessed for enrichment of gene ontology terms. P-value was corrected for multiple comparisons using the Bonferroni method.
| Up-regulated genes |
||
|---|---|---|
| GO term | Fold enrichment | Corrected p-value |
| Monooxygenase activity | 2.09 | 1.10E-08 |
| Metabolic process | 1.29 | 5.50E-07 |
| Electron transport | 1.52 | 7.11E-07 |
| Integral to membrane | 1.44 | 1.31E-06 |
| Oxidoreductase activity | 1.41 | 2.21E-06 |
| Catalytic activity | 1.28 | 2.11E-05 |
| Carbohydrate metabolic process | 1.55 | 1.14E-03 |
| Heme binding | 1.66 | 2.25E-03 |
| Iron ion binding | 1.66 | 7.17E-03 |
| Hydrolyzing O-glycosyl compounds |
1.68 |
1.12E-02 |
| Down-regulated genes | ||
| GO term | Fold enrichment | Corrected p-value |
| Translation | 2.39 | 1.80E-42 |
| Structural constituent of ribosome | 2.56 | 2.46E-33 |
| Ribosome | 2.45 | 2.70E-32 |
| Aminoacyl-tRNA ligase activity | 2.18 | 1.36E-05 |
| tRNA aminoacylation for protein translation | 2.13 | 2.87E-05 |
| Intracellular | 1.31 | 9.82E-05 |
| RNA binding | 1.72 | 7.50E-03 |
| Small ribosomal subunit | 2.69 | 8.40E-03 |
| 5′-3′ exoribonuclease activity | 2.80 | 1.91E-02 |
| Cytoplasm | 1.37 | 2.47E-02 |
Table 3.
Metabolic pathways that significantly change in response to LaeA overexpression. Genes with metabolic pathway annotations were predicted from the A. pseudoterreus genome. Metabolic pathways with at least 2 annotated genes that are enriched for significantly up- or down-regulated genes.
| Up-regulated pathways | ||||
|---|---|---|---|---|
| Pathway | Fold-change (log2) | Up-regulated | Down-regulated | Quantified |
| Styrene degradation | 3.83 | 2 (100%) | 0 (0%) | 2 |
| C5-branched dibasic acid metabolism | 2.64 | 1 (50%) | 0 (0%) | 2 |
| Cysteine degradation | 2.06 | 2 (67%) | 0 (0%) | 3 |
| Glyoxylate and dicarboxylate metabolism | 1.49 | 3 (60%) | 1 (20%) | 5 |
| Purine metabolism | 1.31 | 8 (67%) | 0 (0%) | 12 |
| Beta-alanine metabolism | 0.97 | 7 (78%) | 1 (11%) | 9 |
| Starch and sucrose metabolism | 0.84 | 2 (50%) | 0 (0%) | 4 |
| Mevalonate pathway | 0.80 | 1 (33%) | 0 (0%) | 3 |
| Galactose metabolism (melibiose) | 0.75 | 5 (83%) | 0 (0%) | 6 |
| Glycolysis/gluconeogenesis | 0.66 | 2 (67%) | 0 (0%) | 3 |
| Xenobiotics biodegradation and metabolism | 0.59 | 3 (43%) | 0 (0%) | 7 |
| Stachyose, raffinose, sucrose degradation | 0.58 | 5 (71%) | 0 (0%) | 7 |
| Metabolism of terpenoids and polyketides | 0.55 | 3 (75%) | 1 (25%) | 4 |
| Phenylalanine metabolism | 0.54 | 3 (50%) | 1 (17%) | 6 |
| Starch and sucrose metabolism |
0.51 |
21 (53%) |
7 (18%) |
40 |
| Down-regulated pathways | ||||
| Pathway | Fold-change (log2) | Up-regulated | Down-regulated | Quantified |
| Amino acid metabolism | −0.51 | 0 (0%) | 2 (100%) | 2 |
| Vitamin B6 metabolism | −0.53 | 0 (0%) | 4 (67%) | 6 |
| One carbon pool by folate | −0.62 | 0 (0%) | 4 (100%) | 4 |
| Valine, leucine and isoleucine biosynthesis | −0.68 | 0 (0%) | 3 (100%) | 3 |
| Formation of unsaturated cytosolic fatty acids | −0.74 | 0 (0%) | 3 (75%) | 4 |
| Purine metabolism (salvage pathways) | −0.75 | 0 (0%) | 2 (100%) | 2 |
| Fructose and mannose metabolism | −0.77 | 0 (0%) | 2 (100%) | 2 |
| Biosynthesis of unsaturated fatty acids | −0.77 | 0 (0%) | 1 (50%) | 2 |
| Vanillate degradation | −0.78 | 0 (0%) | 1 (50%) | 2 |
| Amino sugar and nucleotide sugar metabolism | −0.86 | 0 (0%) | 2 (677%) | 3 |
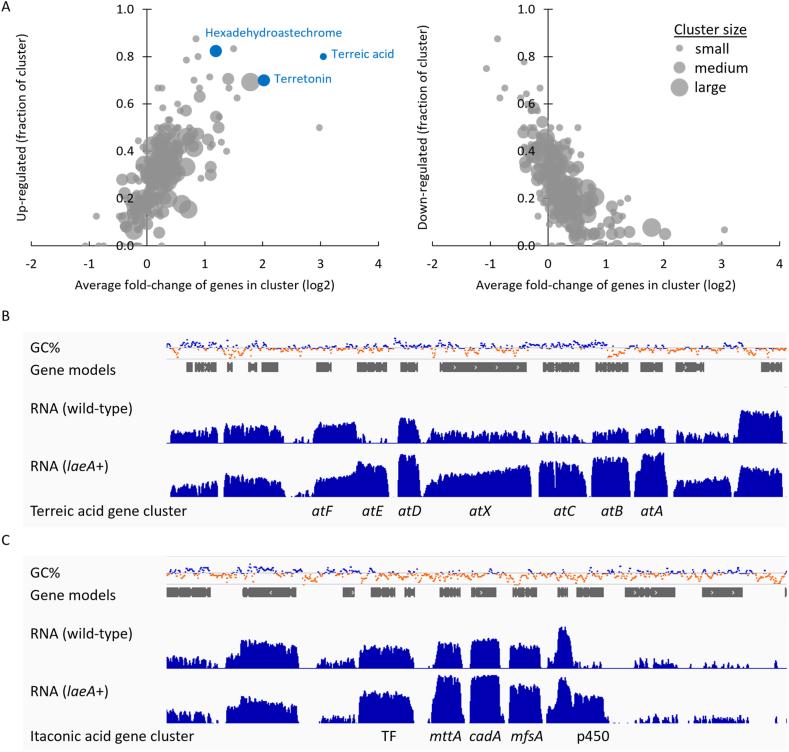
Genes involved in secondary metabolism are not well annotated in the metabolic model. To examine these, we predicted 222 candidate secondary metabolite gene clusters from the genome of A. pseudoterreus using fungiSMASH v4.2.0 (Blin et al., 2017). Genes predicted to occur in secondary metabolite clusters are significantly up-regulated by LaeA overexpression (Fig. 3a). Of these, clusters encoding genes for the biosynthesis of terreic acid (Guo et al., 2014), terretonin (Guo et al., 2012), and hexadehydroastechrome (Yin et al., 2013) are strongly induced (Fig. 3a). It is interesting to note that the transcription factor in the terreic acid cluster (atF) is not significantly altered in expression level, while the biosynthetic genes (atA, atC, atD, atE, and atX) and the transporter (atB) are all significantly up-regulated from 27- to 250-fold (Fig. 3b). The itaconic acid biosynthetic cluster, while not predicted as such, is also induced by overexpression of LaeA (Fig. 3C). We predicted 504 genes encoding carbohydrate active enzymes with Hotpep (Busk et al., 2017), 341 of which were quantified by RNA-seq. The glycoside hydrolase, carbohydrate esterase, carbohydrate binding, and auxiliary activity families tended to have the most genes that were significantly altered in their expression level and are all enriched for genes that are significantly up-regulated by overexpression of LaeA, whereas the glycosyl transferase and polysaccharide lyase families are less impacted (Table 4).
Fig. 3.
Regulation of secondary metabolite clusters by LaeA. The expression level of predicted secondary metabolite clusters was determined in wild-type and LaeA overexpression strains during growth stage in shake flasks. A) The average change in expression of genes within each cluster and the fraction of genes within the cluster that are significantly up- or down-regulated by LaeA overexpression (q < 0.05). Strongly induced clusters with known products are indicated in blue. B) Genomic view of transcriptome data for the terreic acid biosynthetic cluster. C) Genomic view of transcriptome data for the itaconic acid biosynthetic cluster. Transcriptome sequencing depth is shown in log scale. (For interpretation of the references to color in this figure legend, the reader is referred to the Web version of this article.)
Table 4.
Carbohydrate active enzymes that significantly change in response to LaeA overexpression. Carbohydrate-active enzymes were predicted from the A. pseudoterreus genome. Significantly up- and down-regulated genes (q < 0.01) were identified and counted for each family.
| Family | Up-regulated | Down-regulated | Quantified |
|---|---|---|---|
| Glycoside hydrolase | 75 (43%) | 23 (13%) | 173 |
| Glycosyl transferase | 13 (20%) | 13 (20%) | 65 |
| Auxiliary activity | 14 (37%) | 8 (21%) | 38 |
| Carbohydrate binding | 11 (37%) | 4 (13%) | 30 |
| Carbohydrate esterase | 9 (39%) | 3 (13%) | 23 |
| Polysaccharide lyase | 3 (25%) | 2 (17%) | 12 |
3.4. Metabolic analysis of LaeA overexpression
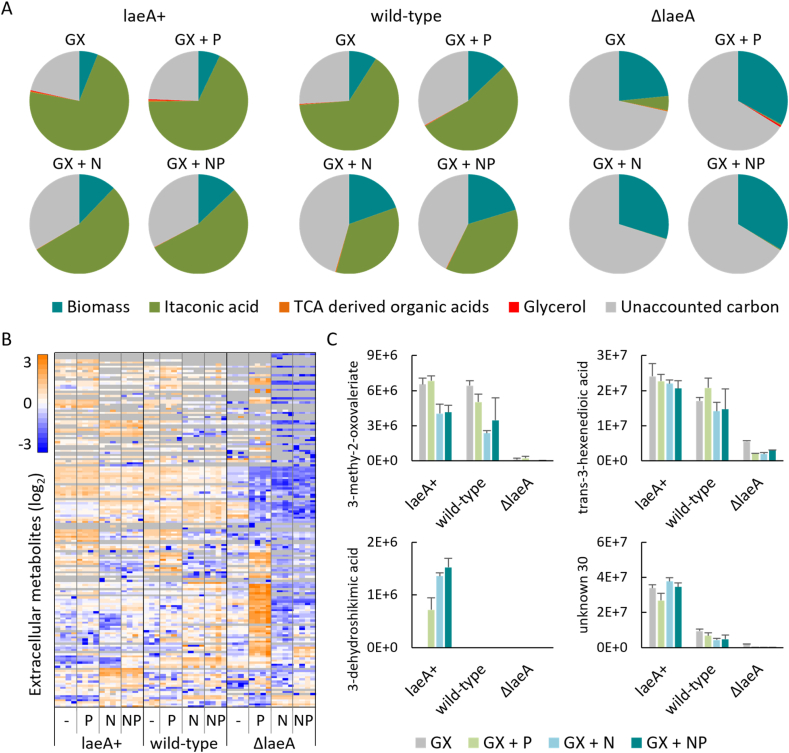
Production of itaconic acid in the medium used in these experiments is dependent on limitation of phosphate (Riscaldati et al., 2000). When the phosphate level was increased (from 0.11 to 0.25 g/L KH2PO4) the wild-type strain produced more biomass and less itaconic acid. In the LaeA overexpression strain the impact of additional phosphate was attenuated while in the deletion strain addition of phosphate decreased itaconic acid production to below the limit of detection (Fig. 2A). We examined the overall carbon balance during itaconic acid production that are controlled by laeA. Biomass (assumed composition of CH1.8O0.5N0.2P0.02) and itaconic acid production accounts for the majority of the carbon from glucose and xylose while the sum of the other products that were absolutely quantified (isocitric acid, cis-aconitic acid, malic acid, succinic acid, citric acid, and glycerol) account for less than 1% of the carbon produced in any of the strains or conditions (Fig. 4a). While biomass production increases dramatically when laeA is deleted, this does not fully account for the loss of itaconic acid production and leaves well over half the carbon unaccounted for in the ΔlaeA strain.
Fig. 4.
Extracellular metabolomics. A) Carbon balance after consumption of 50 g/L glucose and xylose (GX) with addition of phosphate (P) or yeast extract (N) to the medium. B) Global extracellular metabolomics analysis detected 184 metabolites. Hierarchical clustering used the average agglomeration method with the correlation distance metric. C) Relative levels (GC-MS peak area) of selected laeA responsive metabolites.
We performed global extracellular metabolomics analysis to identify products secreted by A. pseudoterreus during itaconic acid production that, in addition to CO2, may account for some of the missing carbon (Fig. 4B). 184 extracellular metabolites were detected (Additional File 1), 103 of which are present at a level higher than that of the control medium in at least one strain. Of these, 31 metabolites were identified while most remain unknown. In general, the level of secreted metabolites in the ΔlaeA strain was lower and no metabolites were quantified that are specifically enriched in the ΔlaeA strain suggesting the unaccounted carbon is primarily in the form of biomass and gaseous products (primarily CO2).
Deletion of laeA resulted in loss of many extracellular metabolites including aromatic compounds such as 3-methoxyanthranilic acid, 3-dehydroxyshikimic acid, and protocatechuic acid as well as a wide variety of unidentified metabolites that may be byproducts of secondary metabolism. Expression of many of these metabolites was increased by laeA overexpression, consistent with control of their production as secondary metabolites. In general, the level of secreted metabolites in the laeA deletion strain was lower, however, addition of phosphate lead to a profound impact on the secreted metabolome (Fig. 3B) that includes higher extracellular concentration of sugars and their derivatives (melibiose, cellobiose, sophorose, lactose, lactulose, lactobionic acid, 1,5-anhydrohexitol, and trehalose), sugar alcohols (xylitol, galactitol, erythropentitol, and palatinitol), and 2,3-butanediol suggesting laeA may be required for sensing or appropriately responding to phosphate availability.
3.5. Proteome analysis of LaeA overexpression
We profiled the proteome of the wild-type and LaeA overexpression strains at three time-points during growth (36h) and transition to phosphate depletion induced organic acid production phase (60h and 84h) to better understand how LaeA regulates the physiology of A. pseudoterreus. Global profiling of wild-type and LaeA overexpression strains of A. pseudoterreus quantified 27,203 peptides (corresponding to 3193 proteins), and 188 targeted peptides were used to quantify 74 proteins involved in central carbon metabolism. Of the quantified proteins, 890 are expressed at a significantly different level between the wild-type and LaeA overexpression strain during at least one time-point (p < 0.05).
To examine the function of the proteins regulated by LaeA, functional enrichment analysis for metabolic pathways, biological processes, molecular functions, and cellular compartments was performed for up- and down-regulated genes at 36, 60, and 84 hours (Table 5). The most significantly enriched (p < 0.05 after Bonferroni correction) gene ontology terms are associated with proteins up- and down-regulated at 84 hours and include electron transport, terpene and polyketide metabolism (up-regulated) and riboflavin, folate, and metabolism of other cofactors and vitamins (down-regulated).
Table 5.
Gene ontology term enrichment in LaeA overexpression strain proteome. Significantly up- and down-regulated proteins in the LaeA overexpression strain versus wild-type during growth (36h) and the transition to production phase (60h and 84h) were assessed for enrichment of gene ontology terms. Hypergeometric test p-values are shown.
| Up-regulated proteins | ||||
|---|---|---|---|---|
| GO term | Hour | Proteins (sig./background) | Fold enrichment | p-value |
| Oxidoreductase activity | 36 | 14/203 | 2.62 | 9.14E-04 |
| One-carbon compound metabolic process | 36 | 2/2 | 28.13 | 1.19E-03 |
| Peroxidase activity | 36 | 3/9 | 14.56 | 1.53E-03 |
| Phospholipase D activity | 36 | 2/3 | 26.85 | 2.25E-03 |
| Protein transporter activity | 36 | 4/21 | 8.29 | 2.28E-03 |
| Metallopeptidase activity | 36 | 3/12 | 11.26 | 3.78E-03 |
| Response to oxidative stress | 36 | 3/12 | 9.02 | 7.00E-03 |
| O-glycosyl compound hydrolase activity | 60 | 4/35 | 8.26 | 2.47E-03 |
| Carbohydrate metabolic process | 60 | 5/67 | 4.71 | 6.82E-03 |
| Serine carboxypeptidase activity | 60 | 2/8 | 20.04 | 7.31E-03 |
| Ubiquinol-cytochrome-c reductase activity | 84 | 3/6 | 20.58 | 3.87E-04 |
| Mitochondrial electron transport | 84 | 2/2 | 34.35 | 7.85E-04 |
| Metabolism of terpenoids and polyketides | 84 | 2/3 | 25.28 | 2.19E-03 |
| Metabolism of other amino acids |
84 |
3/13 |
9.86 |
4.41E-03 |
| Down-regulated proteins | ||||
| GO term | Hour | Proteins (sig./background) | Fold enrichment | p-value |
| Small subunit processome | 36 | 3/4 | 12.57 | 8.92E-04 |
| Protein serine/threonine kinase activity | 60 | 8/38 | 4.38 | 6.39E-04 |
| Hydrolase activity, acting on ester bonds | 60 | 4/9 | 9.51 | 7.79E-04 |
| Metal ion binding | 60 | 6/24 | 5.33 | 1.22E-03 |
| Protein kinase activity | 60 | 8/42 | 3.97 | 1.29E-03 |
| Protein amino acid phosphorylation | 60 | 8/45 | 3.73 | 1.87E-03 |
| Protein-tyrosine kinase activity | 60 | 7/37 | 4.01 | 2.73E-03 |
| Hydrolase activity | 60 | 10/73 | 2.81 | 4.41E-03 |
| Myosin complex | 60 | 2/4 | 15.33 | 8.30E-03 |
| Methyltransferase activity | 60 | 4/16 | 5.65 | 8.42E-03 |
| Riboflavin metabolism | 84 | 6/10 | 7.78 | 3.33E-05 |
| Folate biosynthesis | 84 | 7/15 | 6.13 | 5.65E-05 |
| Metabolism of cofactors and Vitamins | 84 | 13/63 | 2.68 | 6.19E-04 |
| Hydrolase activity, acting on ester bonds | 84 | 4/9 | 8.35 | 1.29E-03 |
| Acid phosphatase activity | 84 | 4/10 | 7.61 | 2.04E-03 |
| Cellular metabolic process | 84 | 5/16 | 5.79 | 2.06E-03 |
| Fatty-acid ligase activity | 84 | 2/2 | 16.31 | 3.66E-03 |
| Double-strand break repair | 84 | 2/2 | 15.98 | 3.78E-03 |
| Fatty acid metabolism | 84 | 2/2 | 12.07 | 6.40E-03 |
| G-protein coupled receptor protein signaling | 84 | 3/7 | 8.23 | 6.68E-03 |
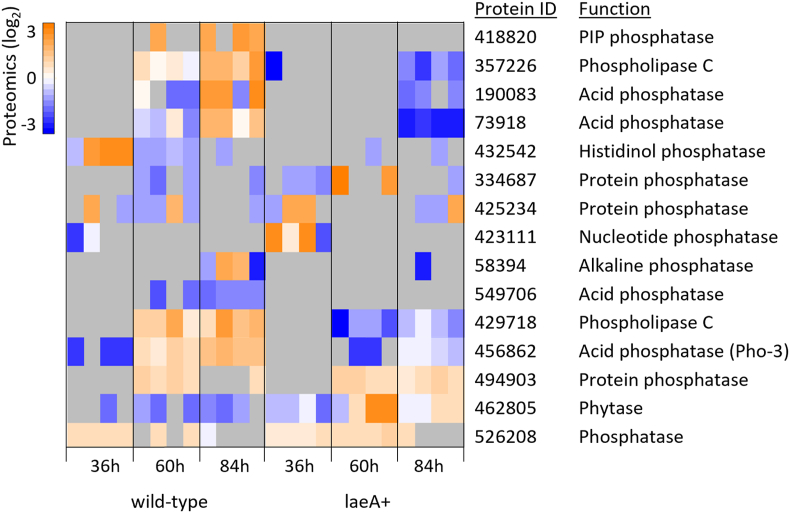
We identified regulatory genes to better understand how LaeA impacts gene regulation in A. pseudoterreus. Kinases, phosphatases, and transcription factors were predicted from the genome of A. pseudoterreus. InterProScan (Jones et al., 2014) was used to identify proteins with kinase or phosphatase domains while transcription factors were identified by homology with known fungal transcription factors present in the Fungal Transcription Factor Database (Park et al., 2008) and the Transcription factor prediction database (Wilson et al., 2008). From this we identified 326 kinases, 142 phosphatases, and 550 transcription factor candidates from the A. pseudoterreus genome, many of which are differentially regulated. Of these proteins the acid phosphatase Pho-3 is the most significantly up-regulated in response to phosphate limitation in the wild-type strain (30.2x from 36h to 84h) and is significantly up-regulated compared with the LaeA overexpression strain. Analysis of all differently expressed phosphatases suggests a much more limited response to phosphate limitation when LaeA is overexpressed (Fig. 5).
Fig. 5.
Differently expressed phosphatases. Phosphatases significantly differentially expressed between the wild-type and LaeA overexpression strain during at least one time-point are shown. Expression of acid phosphatases is attenuated by overexpression of LaeA. Gray boxes indicate proteins below the limit of detection.
4. Discussion and conclusions
Many bioprocesses are optimized around limitation of specific nutrients. Bioreactor conditions to cultivate strains of A. terreus and A. pseudoterreus have been optimized to produce high titers of itaconic acid but often exhibit trade-offs between production rate and overall yield in response to phosphate level (Hevekerl et al., 2014a; Krull et al., 2017; Riscaldati et al., 2000). A low phosphate level is used to limit growth and increase yield and may also limit the impact of excess manganese (Saha and Kennedy, 2020). However, with excess phosphate more biomass is produced allowing a higher overall conversion rate (Hevekerl et al., 2014b). Process control by nutrient limitation is possible under tightly controlled conditions, however heterogeneous feedstocks such as lignocellulosic sugar streams can dramatically impact productivity. In A. pseudoterreus we found that production of organic acids is reduced when using sugars from lignocellulosic feedstocks which contain a mixture of chemicals that act as nutrients and growth inhibitors (Chen et al., 2016). We therefore sought a means to increase the reliability of bioproduction in the absence of specific nutrient limitations. In many fungi the global regulator LaeA controls secondary metabolism and we hypothesized that the itaconic acid gene cluster in A. pseudoterreus (Deng et al., 2020) is controlled by LaeA.
We found that laeA is required for high production of itaconic acid and that overexpression makes itaconic acid production from a mixed glucose/xylose sugar stream more robust to the presence of excess phosphate or mixed nutrients from yeast extract (Fig. 2). Systematic examination of the effects of LaeA overexpression in phosphate limited medium found that proteins with acid phosphatase activity tend to be down-regulated and that genes involved in purine metabolism as well as amino sugar and nucleotide metabolism pathways are significantly down-regulated at the transcript level (Table 3, Table 5) suggesting the response to phosphate limitation and initiation of phosphate scavenging may be delayed or suppressed by overexpression of LaeA.
Metabolic pathways involved in phosphate acquisition are normally repressed but become active in phosphate limited conditions. For example, genetic screens in the filamentous fungus Neurospora crassa and the yeast Saccharomyces cerevisiae have identified a variety of acid and alkaline phosphates, as well as phosphate transport systems that are expressed specifically under conditions of phosphate limitation (Mann et al., 1989; Nelson et al., 1976; Gleason and Metzenberg, 1974). This is controlled by the Pho80/Pho85 cyclin/cyclin dependent kinase complex which phosphorylates the basic helix-loop-helix transcription factor Pho4 (Kaffman et al., 1994). Pho4 then binds upstream of phosphate acquisition genes to regulate their transcription (Peleg and Metzenberg, 1994) while the ankyrin repeat protein Pho81 inhibits phosphorylation by Pho80/Pho85 (Gras et al., 2009; Schneider et al., 1994; Waters et al., 2004). In A. pseudoterreus, when LaeA is overexpressed, 9/13 quantified genes involved in phosphate acquisition are repressed at 36 hours (g < 0.05) and for genes where protein was quantified, such as homologs of the acid phosphatases pho-3 (456,862) and pacA (430,438), this repression is maintained later in the culture at 60 and 84 hours (Table 6), suggesting LaeA overexpression may inhibit activation of these genes in phosphate limited conditions. Of the proteins regulating phosphate acquisition, only the PHO80 cyclin homolog is significantly down-regulated while homologs of PHO4, PHO81, and PHO85 are not affected at 36h.
Table 6.
Genes involved in phosphate sensing and acquisition.A. pseudoterreus genes with homology to genes known to be involved in sensing and regulation of phosphate acquisition in Aspergillus nidulans, Neurospora crassa, and Saccharomyces cerevisiae were identified by BlastP. Delta values indicate the log2 fold-change for the LaeA overexpression strain versus wild-type. Dashes indicate genes that were not detected at the transcript or protein level or for which insufficient data was available for statistical analysis.
| Regulators of phosphate acquisition |
Homologs |
RNA (36h) |
Protein (36h) |
Protein (60h) |
Protein (84h) |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein ID | Annotation | A. nidulans | N. crassa | S. cerevisiae | Δ | g-value | Δ | p-value | Δ | p-value | Δ | p-value |
| 520908 | Ankyrin repeat protein; inhibits Pho80/Pho85 complex | AN4310 | nuc-2 | PHO81 | −0.19 | 6.8E-02 | – | – | – | – | – | – |
| 508770 | Cyclin involved in phosphate homeostasis; interacts with Pho85 | AN5156 | preg | PHO80 | −0.41 | 5.0E-04 | – | – | – | – | – | – |
| 187867 | Cyclin-dependent protein kinase; interacts with cyclin Pho80 | AN8261 | mdk-1 | PHO85 | 0.16 | 1.3E-01 | – | – | – | – | – | – |
| 414825 |
Transcription factor, regulates phosphate acquisition |
AN8271 |
nuc-1 |
PHO4 |
−0.13 |
2.3E-01 |
– |
– |
– |
– |
– |
– |
| Enzymes involved in phosphate acquisition | Homologs | RNA (36h) | Protein (36h) | Protein (60h) | Protein (84h) | |||||||
| Protein ID |
Annotation |
A. nidulans |
N. crassa |
S. cerevisiae |
Δ |
g-value |
Δ |
p-value |
Δ |
p-value |
Δ |
p-value |
| 482564 | Acid phosphatase | AN8063 | pho-13 | – | 0.25 | 1.5E-02 | 1.03 | 1.2E-01 | −0.80 | 5.2E-01 | 1.14 | 8.9E-01 |
| 456862 | Acid phosphatase | AN2360 | pho-3 | – | −1.02 | 1.3E-04 | – | – | −3.68 | 9.2E-05 | −1.90 | 2.8E-05 |
| 7506 | Acid phosphatase | AN4055 | pho-9 | – | −0.16 | 2.2E-01 | – | – | −0.48 | 2.1E-02 | 0.25 | 6.5E-01 |
| 526208 | Acid phosphatase | AN0952 | pho-8 | – | −0.52 | 1.3E-04 | −0.82 | 1.9E-02 | −0.04 | 2.4E-01 | – | – |
| 430438 | Acid phosphatase | AN7142 | – | – | −1.24 | 1.3E-04 | – | – | −2.02 | 1.8E-04 | −1.58 | 1.2E-04 |
| 472595 | Alkaline phosphatase | AN11069 | pho-11 | – | 0.10 | 3.6E-01 | – | – | – | – | – | – |
| 58394 | Alkaline phosphatase | AN2493 | pho-2 | – | −0.39 | 1.2E-02 | – | – | – | – | – | – |
| 456715 | Alkaline phosphatase | AN8622 | pho-12 | – | −0.94 | – | – | – | – | – | – | – |
| 508770 | Vacuolar alkaline phosphatase | AN10563 | pho-10 | PHO8 | −0.40 | 1.3E-04 | – | – | – | – | – | – |
| 523629 | High-affinity phosphate permease | AN0217 | pho-5 | PHO84 | −1.78 | 1.3E-04 | – | – | – | – | – | – |
| 455719 | Low-affinity phosphate permease | AN0469 | pho-6 | PHO91 | −0.39 | 1.3E-04 | – | – | – | – | – | – |
| 203804 | Low-affinity phosphate transporter | AN5935 | pho-7 | PHO84 | −0.55 | 1.3E-04 | – | – | – | – | −1.55 | 5.2E-02 |
| 455671 | Phosphate permease | AN10343 | pho-4 | PHO89 | −0.79 | 1.9E-02 | – | – | – | – | – | – |
| 499196 | Phosphate transporter | AN8040 | pho-15 | PHO88 | −0.15 | 1.4E-01 | – | – | – | – | – | – |
The presence of growth promoting nutrients, such as those found in the complex micro- and macro-nutrient source yeast extract, also significantly decreases production of itaconic acid and pushes the carbon balance toward growth. Like with excess phosphate, overexpression of LaeA limited growth and improved the yield of itaconic acid from mixed sugars when challenged with excess nutrients that may be available in inexpensive lignocellulosic feedstocks or other carbohydrate containing waste-streams such as stillage produced by bioethanol refineries (Kim et al., 2008) or wastewater from potato processing (Lasik et al., 2010).
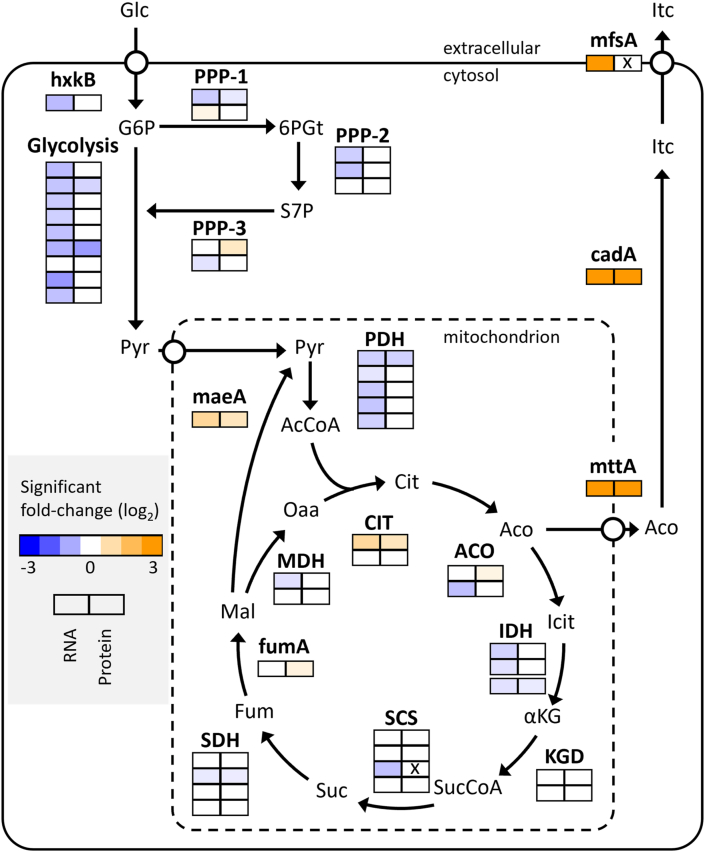
Overexpression of LaeA results in global impacts on cellular functions and metabolism confirming its role as a master regulator in A. pseudoterreus. Notably genes systematically categorized as involved in secondary metabolism were up-regulated in response to LaeA overexpression as has been described in a wide variety of Aspergilli and related fungi (Zhang et al., 2020; Lan et al., 2020; Feng et al., 2020; Grau et al., 2019; Wang et al., 2018; Liu et al., 2016; Jiang et al., 2016; Hong et al., 2015; Oda et al., 2011; Kale et al., 2008; Bok and Keller, 2004). Only more recently has the involvement of laeA in central metabolism been noted for its impact on citric acid production (Linde et al., 2016; Niu et al., 2015). Studies in Aspergillus luchuensis linked the impact of laeA on citric acid production to expression of the citric acid exporter cexA (Kadooka et al., 2020). Likewise, here we found that laeA regulates all of the genes in the itaconic acid cluster including the biosynthetic gene cadA and the mitochondrial and plasma membrane transporters mttA and mfsA (Deng et al., 2020). Within central metabolism, LaeA overexpression resulted in down-regulation of enzymes involved in glycolysis, the pentose phosphate pathway, and formation of acetyl-CoA via the pyruvate dehydrogenase complex but up-regulation of malic enzyme and citrate synthase (Fig. 6). The carbon-balance suggests the ΔlaeA strain produces substantially more CO2 than the strains that produce itaconic acid (Fig. 4), which is notable because CadA itself releases a mole of CO2 for every mole of itaconic acid produced. This suggests that interrupting export of cis-aconitate from the mitochondria promotes continued and potentially futile respiration via cycling of the tricarboxylic acid cycle which produces 2 mol of CO2.
Fig. 6.
Impact of LaeA overexpression on Itaconic acid production. Significant changes in RNA and protein expression level at 36h in shake flasks. Non-significant changes are colored white. None-measured RNA and protein levels are indicated by an ‘X’.
The results presented herein suggest that the higher yield of itaconic acid observed in the LaeA overexpression strain is due not only to increased expression of enzymes in the itaconic acid gene cluster, but decreased growth due to phosphate limitation and increased expression of key enzymes supplying precursor metabolites. Utilization of a global gene regulator such as LaeA to control pathway expression has promise for bioconversion of feedstocks with heterogeneous composition. However, the functionality of LaeA, including its enzymatic activity and target(s) remains an enigma, and increased expression of other secondary metabolite clusters may contaminate the bioproduct with additional complex chemicals and add cost to the purification process.
Availability of data and materials
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Author statement
Kyle R Pomraning: Conceptualization, Methodology, Writing - Original Draft, Writing - Review & Editing, Visualization, Investigation Ziyu Dai: Conceptualization, Visualization, Writing - Review & Editing, Investigation, Nathalie Munoz: Investigation, Young-Mo Kim: Investigation, Yuqian Gao: Investigation, Shuang Deng: Conceptualization, Investigation, Teresa Lemmon: Investigation, Marie S Swita: Investigation, Jeremy D Zucker: Investigation, Joonhoon Kim: Investigation, Stephen J Mondo: Investigation, Ellen Panisko: Investigation, Meagan C Burnet: Investigation, Bobbie-Jo M Webb-Robertson: Formal analysis, Beth Hofstad: Project administration, Investigation, Scott E Baker: Conceptualization, Investigation, Kristin E Burnum-Johnson: Conceptualization, Investigation, Jon K Magnuson: Conceptualization, Project administration, Funding acquisition.
Funding
The research was supported by the U.S. Department of Energy (DOE), Office of Energy Efficiency and Renewable Energy (EERE), Bioenergy Technologies Office (BETO), under Award No. DE-NL0030038. The multi-omic analysis in the current research was performed using EMSL (grid.436923.9), a DOE Office of Science User Facility sponsored by the Office of Biological and Environmental Research. Pacific Northwest National Laboratory is multi-program national laboratory operated by Battelle for the DOE under Contract No. DE-AC05-76RLO 1830. The work conducted by the U.S. Department of Energy Joint Genome Institute (https://ror.org/04xm1d337), a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy operated under Contract No.DE-AC02-05CH11231. The views expressed in the article do not necessarily represent the views of the U.S. Department of Energy or the United States Government.
Additional Files
Additional file 1. xlsx
Excel file containing RNA-seq analysis as well as targeted and global proteomics data and extracellular metabolomics data.
Declaration of competing interest
The authors declare no competing interests.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.mec.2022.e00203.
Contributor Information
Kyle R. Pomraning, Email: Kyle.Pomraning@pnnl.gov.
Ziyu Dai, Email: Ziyu.Dai@pnnl.gov.
Nathalie Munoz, Email: Nathalie.munozmunoz@pnnl.gov.
Young-Mo Kim, Email: Young-Mo.Kim@pnnl.gov.
Yuqian Gao, Email: Yuqian.Gao@pnnl.gov.
Shuang Deng, Email: Shuang.Deng@pnnl.gov.
Teresa Lemmon, Email: Teresa.Lemmon@pnnl.gov.
Marie S. Swita, Email: Marie.Swita@pnnl.gov.
Jeremy D. Zucker, Email: Jeremy.Zucker@pnnl.gov.
Joonhoon Kim, Email: Joonhoon.Kim@pnnl.gov.
Stephen J. Mondo, Email: sjmondo@lbl.gov.
Ellen Panisko, Email: Ellen.Panisko@pnnl.gov.
Meagan C. Burnet, Email: Meagan.Burnet@pnnl.gov.
Bobbie-Jo M. Webb-Robertson, Email: Bobbie-Jo.Webb-Robertson@pnnl.gov.
Beth Hofstad, Email: Beth.Hofstad@pnnl.gov.
Scott E. Baker, Email: Scott.Baker@pnnl.gov.
Kristin E. Burnum-Johnson, Email: kristin.burnum-johnson@pnnl.gov.
Jon K. Magnuson, Email: Jon.Magnuson@pnnl.gov.
List of abbreviations
- MM –
minimal medium
- PM
production medium
- GX
minimal glucose/xylose medium
- GX + P
minimal glucose/xylose medium with added phosphate
- GX + N
minimal glucose/xylose medium with added yeast extract
- GX + NP
minimal glucose/xylose medium with added yeast extract and phosphate
- GM
germination medium
- DMR
medium with sugars from deacetylated and mechanically refined corn stover
- atA
terreic acid cluster gene A
- afB
terreic acid cluster gene B
- atC
terreic acid cluster gene C
- atD
terreic acid cluster gene D
- atE
terreic acid cluster gene E
- atF
terreic acid cluster gene F
- TF
itaconic acid gene cluster zinc-finger transcription factor
- mttA
itaconic acid gene cluster mitochondrial transporter
- cad
itaconic acid gene cluster cis-aconitate decarboxylase
- mfsA
itaconic acid gene cluster major facilitator superfamily transporter
- p450
itaconic acid gene cluster p450 domain containing protein
Appendix A. Supplementary data
The following is/are the supplementary data to this article:
References
- Amare M.G., Keller N.P. Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 2014;66:11–18. doi: 10.1016/j.fgb.2014.02.008. [DOI] [PubMed] [Google Scholar]
- Bafana R., Pandey R.A. New approaches for itaconic acid production: bottlenecks and possible remedies. Crit. Rev. Biotechnol. 2018;38(1):68–82. doi: 10.1080/07388551.2017.1312268. [DOI] [PubMed] [Google Scholar]
- Bayram O., Braus G.H. Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol. Rev. 2012;36(1):1–24. doi: 10.1111/j.1574-6976.2011.00285.x. [DOI] [PubMed] [Google Scholar]
- Bayram O., Krappmann S., Ni M., Bok J.W., Helmstaedt K., Valerius O., Braus-Stromeyer S., Kwon N.J., Keller N.P., Yu J.H., et al. VelB/VeA/LaeA complex coordinates light signal with fungal development and secondary metabolism. Science. 2008;320(5882):1504–1506. doi: 10.1126/science.1155888. [DOI] [PubMed] [Google Scholar]
- Becker J., Hosseinpour Tehrani H., Gauert M., Mampel J., Blank L.M., Wierckx N. An Ustilago maydis chassis for itaconic acid production without by-products. Microb. Biotechnol. 2020;13(2):350–362. doi: 10.1111/1751-7915.13525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blin K., Wolf T., Chevrette M.G., Lu X., Schwalen C.J., Kautsar S.A., Suarez Duran H.G., de Los Santos E.L.C., Kim H.U., Nave M., et al. antiSMASH 4.0-improvements in chemistry prediction and gene cluster boundary identification. Nucleic Acids Res. 2017;45(W1):W36–W41. doi: 10.1093/nar/gkx319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bok J.W., Keller N.P. LaeA, a regulator of secondary metabolism in Aspergillus spp. Eukaryot. Cell. 2004;3(2):527–535. doi: 10.1128/EC.3.2.527-535.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busk P.K., Pilgaard B., Lezyk M.J., Meyer A.S., Lange L. Homology to peptide pattern for annotation of carbohydrate-active enzymes and prediction of function. BMC Bioinf. 2017;18(1):214. doi: 10.1186/s12859-017-1625-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callister S.J., Barry R.C., Adkins J.N., Johnson E.T., Qian W.J., Webb-Robertson B.J., Smith R.D., Lipton M.S. Normalization approaches for removing systematic biases associated with mass spectrometry and label-free proteomics. J. Proteome Res. 2006;5(2):277–286. doi: 10.1021/pr050300l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X.W., Wang W., Ciesielski P., Trass O., Park S., Tao L., Tucker M.P. Improving sugar yields and reducing enzyme loadings in the deacetylation and mechanical refining (DMR) process through multistage disk and szego refining and corresponding techno-economic analysis. ACS Sustain. Chem. Eng. 2016;4(1):324–333. [Google Scholar]
- Curtis D.S., Phillips A.R., Callister S.J., Conlan S., McCue L.A. SPOCS: software for predicting and visualizing orthology/paralogy relationships among genomes. Bioinformatics. 2013;29(20):2641–2642. doi: 10.1093/bioinformatics/btt454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Z., Baker S.E. WO2017074533A1. United States: Battelle Memorial Institute; Richland, WA): 2016. (Enhanced Itaconic Acid Production in Aspergillus with Increased Laea Expression). [Google Scholar]
- Dai Z., Aryal U.K., Shukla A., Qian W.J., Smith R.D., Magnuson J.K., Adney W.S., Beckham G.T., Brunecky R., Himmel M.E., et al. Impact of alg3 gene deletion on growth, development, pigment production, protein secretion, and functions of recombinant Trichoderma reesei cellobiohydrolases in Aspergillus Niger. Fungal Genet. Biol. 2013;61:120–132. doi: 10.1016/j.fgb.2013.09.004. [DOI] [PubMed] [Google Scholar]
- Deng S., Dai Z., Swita M., Pomraning K.R., Hofstad B., Panisko E., Baker S., Magnuson J. Deletion analysis of the itaconic acid biosynthesis gene cluster components in Aspergillus pseudoterreus ATCC32359. Appl. Microbiol. Biotechnol. 2020;104(9):3981–3992. doi: 10.1007/s00253-020-10418-0. [DOI] [PubMed] [Google Scholar]
- Eimhjellen K.E., Larsen H. The mechanism of itaconic acid formation by Aspergillus terreus. 2. The effect of substrates and inhibitors. Biochem. J. 1955;60(1):139–147. doi: 10.1042/bj0600139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., Yin Z., Wu Y., Xu L., Du H., Wang N., Huang L. LaeA controls virulence and secondary metabolism in apple canker pathogen valsa Mali. Front. Microbiol. 2020;11 doi: 10.3389/fmicb.2020.581203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerke J., Braus G.H. Manipulation of fungal development as source of novel secondary metabolites for biotechnology. Appl. Microbiol. Biotechnol. 2014;98(20):8443–8455. doi: 10.1007/s00253-014-5997-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson D.G., Young L., Chuang R.Y., Venter J.C., Hutchison C.A., 3rd, Smith H.O. Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat. Methods. 2009;6(5):343–345. doi: 10.1038/nmeth.1318. [DOI] [PubMed] [Google Scholar]
- Gleason M.K., Metzenberg R.L. Regulation of phosphate metabolism in Neurospora crassa: isolation of mutants deficient in ther repressible alkaline phosphatase. Genetics. 1974;78(2):645–659. doi: 10.1093/genetics/78.2.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gras D.E., Silveira H.C., Peres N.T., Sanches P.R., Martinez-Rossi N.M., Rossi A. Transcriptional changes in the nuc-2A mutant strain of Neurospora crassa cultivated under conditions of phosphate shortage. Microbiol. Res. 2009;164(6):658–664. doi: 10.1016/j.micres.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Grau M.F., Entwistle R., Oakley C.E., Wang C.C.C., Oakley B.R. Overexpression of an LaeA-like methyltransferase upregulates secondary metabolite production in Aspergillus nidulans. ACS Chem. Biol. 2019;14(7):1643–1651. doi: 10.1021/acschembio.9b00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigoriev I.V., Nikitin R., Haridas S., Kuo A., Ohm R., Otillar R., Riley R., Salamov A., Zhao X., Korzeniewski F., et al. MycoCosm portal: gearing up for 1000 fungal genomes. Nucleic Acids Res. 2014;42(Database issue):D699–D704. doi: 10.1093/nar/gkt1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.J., Knox B.P., Chiang Y.M., Lo H.C., Sanchez J.F., Lee K.H., Oakley B.R., Bruno K.S., Wang C.C. Molecular genetic characterization of a cluster in A. terreus for biosynthesis of the meroterpenoid terretonin. Org. Lett. 2012;14(22):5684–5687. doi: 10.1021/ol302682z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo C.J., Sun W.W., Bruno K.S., Wang C.C. Molecular genetic characterization of terreic acid pathway in Aspergillus terreus. Org. Lett. 2014;16(20):5250–5253. doi: 10.1021/ol502242a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hevekerl A., Kuenz A., Vorlop K.D. Filamentous fungi in microtiter plates-an easy way to optimize itaconic acid production with Aspergillus terreus. Appl. Microbiol. Biotechnol. 2014;98(16):6983–6989. doi: 10.1007/s00253-014-5743-2. [DOI] [PubMed] [Google Scholar]
- Hevekerl A., Kuenz A., Vorlop K.D. Influence of the pH on the itaconic acid production with Aspergillus terreus. Appl. Microbiol. Biotechnol. 2014;98(24):10005–10012. doi: 10.1007/s00253-014-6047-2. [DOI] [PubMed] [Google Scholar]
- Hong E.J., Kim N.K., Lee D., Kim W.G., Lee I. Overexpression of the laeA gene leads to increased production of cyclopiazonic acid in Aspergillus fumisynnematus. Fungal Biol. 2015;119(11):973–983. doi: 10.1016/j.funbio.2015.06.006. [DOI] [PubMed] [Google Scholar]
- Hosseinpour Tehrani H., Becker J., Bator I., Saur K., Meyer S., Rodrigues Loia A.C., Blank L.M., Wierckx N. Integrated strain- and process design enable production of 220 g L(-1) itaconic acid with Ustilago maydis. Biotechnol. Biofuels. 2019;12:263. doi: 10.1186/s13068-019-1605-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang T., Wang M., Li L., Si J., Song B., Zhou C., Yu M., Wang X., Zhang Y., Ding G., et al. Overexpression of the global regulator LaeA in Chaetomium globosum leads to the biosynthesis of chaetoglobosin Z. J. Nat. Prod. 2016;79(10):2487–2494. doi: 10.1021/acs.jnatprod.6b00333. [DOI] [PubMed] [Google Scholar]
- Jones P., Binns D., Chang H.Y., Fraser M., Li W., McAnulla C., McWilliam H., Maslen J., Mitchell A., Nuka G., et al. InterProScan 5: genome-scale protein function classification. Bioinformatics. 2014;30(9):1236–1240. doi: 10.1093/bioinformatics/btu031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadooka C., Nakamura E., Mori K., Okutsu K., Yoshizaki Y., Takamine K., Goto M., Tamaki H., Futagami T. LaeA controls citric acid production through regulation of the citrate exporter-encoding cexA gene in Aspergillus luchuensis mut. Kawachii. Appl. Environ. Microbiol. 2020;86(5) doi: 10.1128/AEM.01950-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaffman A., Herskowitz I., Tjian R., O'Shea E.K. Phosphorylation of the transcription factor PHO4 by a cyclin-CDK complex, PHO80-PHO85. Science. 1994;263(5150):1153–1156. doi: 10.1126/science.8108735. [DOI] [PubMed] [Google Scholar]
- Kale S.P., Milde L., Trapp M.K., Frisvad J.C., Keller N.P., Bok J.W. Requirement of LaeA for secondary metabolism and sclerotial production in Aspergillus flavus. Fungal Genet. Biol. 2008;45(10):1422–1429. doi: 10.1016/j.fgb.2008.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karaffa L., Diaz R., Papp B., Fekete E., Sandor E., Kubicek C.P. A deficiency of manganese ions in the presence of high sugar concentrations is the critical parameter for achieving high yields of itaconic acid by Aspergillus terreus. Appl. Microbiol. Biotechnol. 2015;99(19):7937–7944. doi: 10.1007/s00253-015-6735-6. [DOI] [PubMed] [Google Scholar]
- Karimi Aghcheh R., Druzhinina I.S., Kubicek C.P. The putative protein methyltransferase LAE1 of Trichoderma atroviride is a key regulator of asexual development and mycoparasitism. PLoS One. 2013;8(6) doi: 10.1371/journal.pone.0067144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi-Aghcheh R., Bok J.W., Phatale P.A., Smith K.M., Baker S.E., Lichius A., Omann M., Zeilinger S., Seiboth B., Rhee C., et al. Functional analyses of Trichoderma reesei LAE1 reveal conserved and contrasting roles of this regulator. G3 (Bethesda) 2013;3(2):369–378. doi: 10.1534/g3.112.005140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y., Mosier N.S., Hendrickson R., Ezeji T., Blaschek H., Dien B., Cotta M., Dale B., Ladisch M.R. Composition of corn dry-grind ethanol by-products: DDGS, wet cake, and thin stillage. Bioresour. Technol. 2008;99(12):5165–5176. doi: 10.1016/j.biortech.2007.09.028. [DOI] [PubMed] [Google Scholar]
- Kollath I.S., Molnar A.P., Soos A., Fekete E., Sandor E., Kovacs B., Kubicek C.P., Karaffa L. Manganese deficiency is required for high itaconic acid production from D-xylose in Aspergillus terreus. Front. Microbiol. 2019;10:1589. doi: 10.3389/fmicb.2019.01589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krull S., Hevekerl A., Kuenz A., Prusse U. Process development of itaconic acid production by a natural wild type strain of Aspergillus terreus to reach industrially relevant final titers. Appl. Microbiol. Biotechnol. 2017;101(10):4063–4072. doi: 10.1007/s00253-017-8192-x. [DOI] [PubMed] [Google Scholar]
- Kuenz A., Krull S. Biotechnological production of itaconic acid-things you have to know. Appl. Microbiol. Biotechnol. 2018;102(9):3901–3914. doi: 10.1007/s00253-018-8895-7. [DOI] [PubMed] [Google Scholar]
- Lan N., Yue Q., An Z., Bills G.F. Apc.LaeA and Apc.VeA of the velvet complex govern secondary metabolism and morphological development in the echinocandin-producing fungus Aspergillus pachycristatus. J. Ind. Microbiol. Biotechnol. 2020;47(1):155–168. doi: 10.1007/s10295-019-02250-x. [DOI] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9(4):357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen H., Eimhjellen K.E. The mechanism of itaconic acid formation by Aspergillus terreus. 1. The effect of acidity. Biochem. J. 1955;60(1):135–139. doi: 10.1042/bj0600135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasik M., Nowak J., Krzywonos M., Cibis E. Impact of batch, repeated-batch (with cell recycle and medium replacement) and continuous processes on the course and efficiency of aerobic thermophilic biodegradation of potato processing wastewater. Bioresour. Technol. 2010;101(10):3444–3451. doi: 10.1016/j.biortech.2009.12.096. [DOI] [PubMed] [Google Scholar]
- Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li A., van Luijk N., ter Beek M., Caspers M., Punt P., van der Werf M. A clone-based transcriptomics approach for the identification of genes relevant for itaconic acid production in Aspergillus. Fungal Genet. Biol. 2011;48(6):602–611. doi: 10.1016/j.fgb.2011.01.013. [DOI] [PubMed] [Google Scholar]
- Linde T., Zoglowek M., Lubeck M., Frisvad J.C., Lubeck P.S. The global regulator LaeA controls production of citric acid and endoglucanases in Aspergillus carbonarius. J. Ind. Microbiol. Biotechnol. 2016;43(8):1139–1147. doi: 10.1007/s10295-016-1781-3. [DOI] [PubMed] [Google Scholar]
- Liu J., Gao Q., Xu N., Liu L. Genome-scale reconstruction and in silico analysis of Aspergillus terreus metabolism. Mol. Biosyst. 2013;9(7):1939–1948. doi: 10.1039/c3mb70090a. [DOI] [PubMed] [Google Scholar]
- Liu Q., Cai L., Shao Y., Zhou Y., Li M., Wang X., Chen F. Inactivation of the global regulator LaeA in Monascus ruber results in a species-dependent response in sporulation and secondary metabolism. Fungal Biol. 2016;120(3):297–305. doi: 10.1016/j.funbio.2015.10.008. [DOI] [PubMed] [Google Scholar]
- Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann B.J., Bowman B.J., Grotelueschen J., Metzenberg R.L. Nucleotide sequence of pho-4+, encoding a phosphate-repressible phosphate permease of Neurospora crassa. Gene. 1989;83(2):281–289. doi: 10.1016/0378-1119(89)90114-5. [DOI] [PubMed] [Google Scholar]
- Matzke M.M., Waters K.M., Metz T.O., Jacobs J.M., Sims A.C., Baric R.S., Pounds J.G., Webb-Robertson B.J. Improved quality control processing of peptide-centric LC-MS proteomics data. Bioinformatics. 2011;27(20):2866–2872. doi: 10.1093/bioinformatics/btr479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matzke M.M., Brown J.N., Gritsenko M.A., Metz T.O., Pounds J.G., Rodland K.D., Shukla A.K., Smith R.D., Waters K.M., McDermott J.E., et al. A comparative analysis of computational approaches to relative protein quantification using peptide peak intensities in label-free LC-MS proteomics experiments. Proteomics. 2013;13(3–4):493–503. doi: 10.1002/pmic.201200269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa M., Nakamura I., Kobayashi T. Process for concentrating and purifying itaconic acid from fermented liquor with electrodialysis. J. Ferment. Technol. 1975;53(5):286–293. [Google Scholar]
- Nelson R.E., Lehman J.F., Metzenberg R.L. Regulation of phosphate metabolism in Neurospora crassa: identification of the structural gene for repressible acid phosphatase. Genetics. 1976;84(2):183–192. doi: 10.1093/genetics/84.2.183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu J., Arentshorst M., Nair P.D., Dai Z., Baker S.E., Frisvad J.C., Nielsen K.F., Punt P.J., Ram A.F. Identification of a classical mutant in the industrial host Aspergillus Niger by systems genetics: LaeA is required for citric acid production and regulates the formation of some secondary metabolites. G3 (Bethesda) 2015;6(1):193–204. doi: 10.1534/g3.115.024067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda K., Kobayashi A., Ohashi S., Sano M. Aspergillus oryzae laeA regulates kojic acid synthesis genes. Biosci. Biotechnol. Biochem. 2011;75(9):1832–1834. doi: 10.1271/bbb.110235. [DOI] [PubMed] [Google Scholar]
- Park J., Park J., Jang S., Kim S., Kong S., Choi J., Ahn K., Kim J., Lee S., Kim S., et al. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008;24(7):1024–1025. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- Pathan M., Keerthikumar S., Ang C.S., Gangoda L., Quek C.Y., Williamson N.A., Mouradov D., Sieber O.M., Simpson R.J., Salim A., et al. FunRich: an open access standalone functional enrichment and interaction network analysis tool. Proteomics. 2015;15(15):2597–2601. doi: 10.1002/pmic.201400515. [DOI] [PubMed] [Google Scholar]
- Peleg Y., Metzenberg R.L. Analysis of the DNA-binding and dimerization activities of Neurospora crassa transcription factor NUC-1. Mol. Cell Biol. 1994;14(12):7816–7826. doi: 10.1128/mcb.14.12.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polpitiya A.D., Qian W.J., Jaitly N., Petyuk V.A., Adkins J.N. Camp DG, 2nd, Anderson GA, Smith RD: DAnTE: a statistical tool for quantitative analysis of -omics data. Bioinformatics. 2008;24(13):1556–1558. doi: 10.1093/bioinformatics/btn217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pomraning K.R., Dai Z., Munoz N., Kim Y.M., Gao Y., Deng S., Kim J., Hofstad B.A., Swita M.S., Lemmon T., et al. Integration of proteomics and metabolomics into the design, build, test, learn cycle to improve 3-hydroxypropionic acid production in Aspergillus pseudoterreus. Front. Bioeng. Biotechnol. 2021;9 doi: 10.3389/fbioe.2021.603832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riscaldati E., Moresi M., Federici F., Petruccioli M. Effect of pH and stirring rate on itaconate production by Aspergillus terreus. J. Biotechnol. 2000;83(3):219–230. doi: 10.1016/s0168-1656(00)00322-9. [DOI] [PubMed] [Google Scholar]
- Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29(1):24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha B.C., Kennedy G.J. Mannose and galactose as substrates for production of itaconic acid by Aspergillus terreus. Lett. Appl. Microbiol. 2017;65(6):527–533. doi: 10.1111/lam.12810. [DOI] [PubMed] [Google Scholar]
- Saha B.C., Kennedy G.J. Ninety six well microtiter plate as microbioreactors for production of itaconic acid by six Aspergillus terreus strains. J. Microbiol. Methods. 2018;144:53–59. doi: 10.1016/j.mimet.2017.11.002. [DOI] [PubMed] [Google Scholar]
- Saha B.C., Kennedy G.J. Efficient itaconic acid production by Aspergillus terreus: overcoming the strong inhibitory effect of manganese. Biotechnol. Prog. 2020;36(2):e2939. doi: 10.1002/btpr.2939. [DOI] [PubMed] [Google Scholar]
- Saha B.C., Kennedy G.J., Qureshi N., Bowman M.J. Production of itaconic acid from pentose sugars by Aspergillus terreus. Biotechnol. Prog. 2017;33(4):1059–1067. doi: 10.1002/btpr.2485. [DOI] [PubMed] [Google Scholar]
- Saha B.C., Kennedy G.J., Bowman M.J., Qureshi N., Dunn R.O. Factors affecting production of itaconic acid from mixed sugars by Aspergillus terreus. Appl. Biochem. Biotechnol. 2019;187(2):449–460. doi: 10.1007/s12010-018-2831-2. [DOI] [PubMed] [Google Scholar]
- Samson R.A., Peterson S.W., Frisvad J.C., Varga J. New species in Aspergillus section Terrei. Stud. Mycol. 2011;69(1):39–55. doi: 10.3114/sim.2011.69.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandor E., Kollath I.S., Fekete E., Biro V., Flipphi M., Kovacs B., Kubicek C.P., Karaffa L. Carbon-source dependent interplay of copper and manganese ions modulates the morphology and itaconic acid production in Aspergillus terreus. Front. Microbiol. 2021;12 doi: 10.3389/fmicb.2021.680420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider K.R., Smith R.L., O'Shea E.K. Phosphate-regulated inactivation of the kinase PHO80-PHO85 by the CDK inhibitor PHO81. Science. 1994;266(5182):122–126. doi: 10.1126/science.7939631. [DOI] [PubMed] [Google Scholar]
- Shin W.S., Park B., Lee D., Oh M.K., Chun G.T., Kim S. Enhanced production of itaconic acid through development of transformed fungal strains of Aspergillus terreus. J. Microbiol. Biotechnol. 2017;27(2):306–315. doi: 10.4014/jmb.1611.11054. [DOI] [PubMed] [Google Scholar]
- Steiger M.G., Punt P.J., Ram A.F.J., Mattanovich D., Sauer M. Characterizing MttA as a mitochondrial cis-aconitic acid transporter by metabolic engineering. Metab. Eng. 2016;35:95–104. doi: 10.1016/j.ymben.2016.02.003. [DOI] [PubMed] [Google Scholar]
- Sun W., Vila-Santa A., Liu N., Prozorov T., Xie D., Faria N.T., Ferreira F.C., Mira N.P., Shao Z. Metabolic engineering of an acid-tolerant yeast strain Pichia kudriavzevii for itaconic acid production. Metab Eng Commun. 2020;10 doi: 10.1016/j.mec.2020.e00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 2021;38(7):3022–3027. doi: 10.1093/molbev/msab120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W T., P G., A A., B J., H J., W J., M A., E D., L L., J S., et al. Office of Biomass Program. EERE); 2004. Top value added chemicals from biomass. [Google Scholar]
- Wang B., Lv Y., Li X., Lin Y., Deng H., Pan L. Profiling of secondary metabolite gene clusters regulated by LaeA in Aspergillus Niger FGSC A1279 based on genome sequencing and transcriptome analysis. Res. Microbiol. 2018;169(2):67–77. doi: 10.1016/j.resmic.2017.10.002. [DOI] [PubMed] [Google Scholar]
- Waters N.C., Knight J.P., Creasy C.L., Bergman L.W. The yeast Pho80-Pho85 cyclin-CDK complex has multiple substrates. Curr. Genet. 2004;46(1):1–9. doi: 10.1007/s00294-004-0501-0. [DOI] [PubMed] [Google Scholar]
- Webb-Robertson B.J., McCue L.A., Waters K.M., Matzke M.M., Jacobs J.M., Metz T.O., Varnum S.M., Pounds J.G. Combined statistical analyses of peptide intensities and peptide occurrences improves identification of significant peptides from MS-based proteomics data. J. Proteome Res. 2010;9(11):5748–5756. doi: 10.1021/pr1005247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb-Robertson B.M., Bramer L.M., Jensen J.L., Kobold M.A., Stratton K.G., White A.M., Rodland K.D. P-MartCancer-Interactive online software to enable analysis of shotgun cancer proteomic datasets. Cancer Res. 2017;77(21):e47–e50. doi: 10.1158/0008-5472.CAN-17-0335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willke T., Vorlop K.D. Biotechnological production of itaconic acid. Appl. Microbiol. Biotechnol. 2001;56(3–4):289–295. doi: 10.1007/s002530100685. [DOI] [PubMed] [Google Scholar]
- Wilson D., Charoensawan V., Kummerfeld S.K., Teichmann S.A. DBD--taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36(Database issue):D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie H., Ma Q., Wei D., Wang F. Metabolic engineering of an industrial Aspergillus Niger strain for itaconic acid production. 3 Biotech. 2020;10(3):113. doi: 10.1007/s13205-020-2080-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin W.B., Baccile J.A., Bok J.W., Chen Y., Keller N.P., Schroeder F.C. A nonribosomal peptide synthetase-derived iron(III) complex from the pathogenic fungus Aspergillus fumigatus. J. Am. Chem. Soc. 2013;135(6):2064–2067. doi: 10.1021/ja311145n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J.H., Hamari Z., Han K.H., Seo J.A., Reyes-Dominguez Y., Scazzocchio C. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 2004;41(11):973–981. doi: 10.1016/j.fgb.2004.08.001. [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang H., Zhu Q., Hao S., Chai S., Li Y., Jiao Z., Shi J., Sun B., Wang C. Overexpression of global regulator LaeA increases secondary metabolite production in Monascus purpureus. Appl. Microbiol. Biotechnol. 2020;104(7):3049–3060. doi: 10.1007/s00253-020-10379-4. [DOI] [PubMed] [Google Scholar]
- Zhao C., Chen S., Fang H. Consolidated bioprocessing of lignocellulosic biomass to itaconic acid by metabolically engineering Neurospora crassa. Appl. Microbiol. Biotechnol. 2018;102(22):9577–9584. doi: 10.1007/s00253-018-9362-1. [DOI] [PubMed] [Google Scholar]
- Zhao C., Cui Z., Zhao X., Zhang J., Zhang L., Tian Y., Qi Q., Liu J. Enhanced itaconic acid production in Yarrowia lipolytica via heterologous expression of a mitochondrial transporter MTT. Appl. Microbiol. Biotechnol. 2019;103(5):2181–2192. doi: 10.1007/s00253-019-09627-z. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated or analyzed during this study are included in this published article and its supplementary information files.