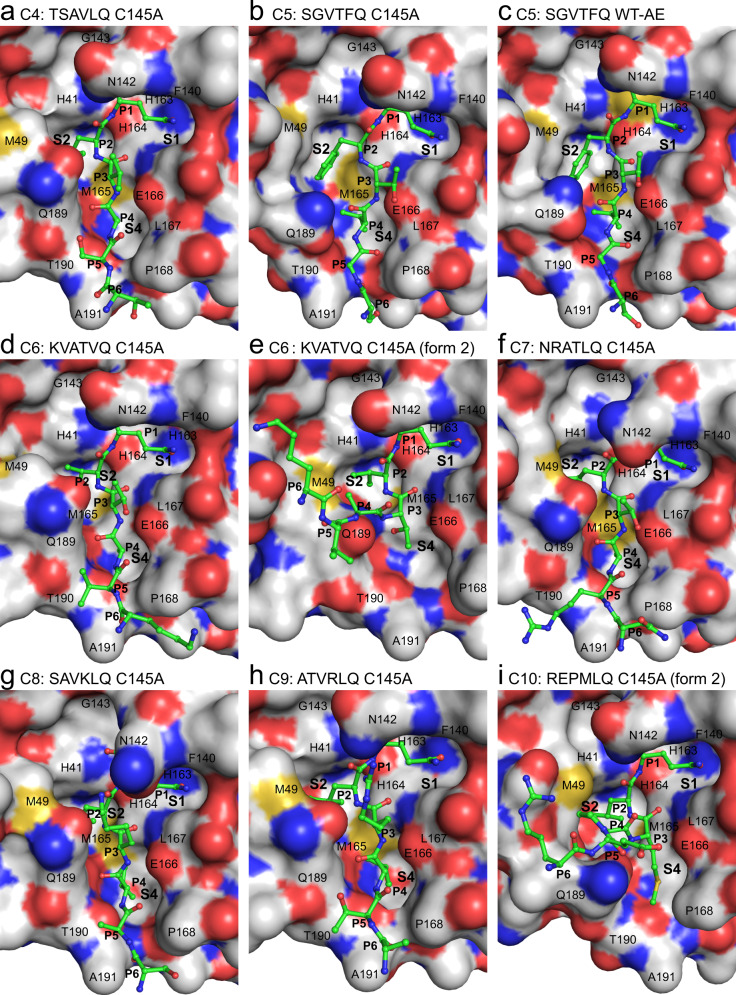
Fig. 2. X-ray crystallographic structures of SARS-CoV-2 Mpro cleavage sites C4 to C10.
a, b and d–i show product-like complexes with the C145A mutant, c shows an acyl-enzyme (AE) intermediate complex with wild-type (WT) Mpro. The complementary substrate specificity residues (P6-P1) are labeled and shown as ball-and-stick (carbons are green) bound into the Mpro substrate binding groove shown as a molecular surface (carbons are gray). The Schechter–Berger substrate specificity pockets (S1, S2, and S4 in Mpro) are labeled. Enzyme residues near the cleavage site atoms are labeled on the molecular surface. Non-carbon atoms are colored as follows: oxygen - red, nitrogen - blue, sulfur - yellow. The C5 C145A (b) and WT (c) structures were determined previously24 with PDB 7JOY and 7KHP, respectively, used to make the figure.