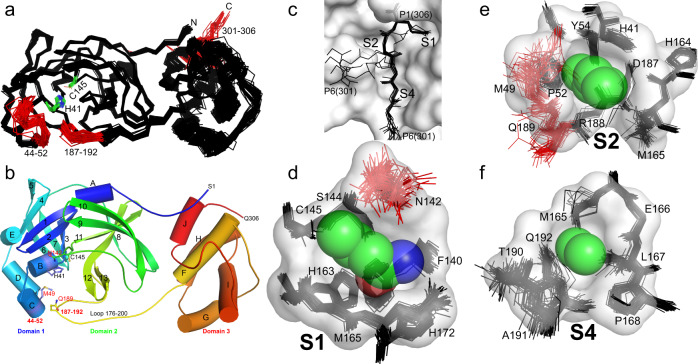
Fig. 4. Plasticity of SARS-CoV-2 Mpro to promote molecular recognition of the polyprotein cleavage site variants.
a Global structural alignment of all unique chains from the multiple structures characterized in this study. Structural alignments were performed using the ALIGN function in PyMOL with all protein atoms. Mobile regions are highlighted in red and labeled. Catalytic dyad H41/C145 are shown with green carbons and labeled. b Mpro cartoon highlighting secondary structural features, oriented as in panel a, colored spectrally - blue N-terminus to red C-terminus. c Overlay of representative P6 to P1 regions observed amongst the distinct cleavage site structures, highlighting their varying main chain conformations. The majority adopt the canonical extended β-type conformation within the binding site groove (see also Figs. 2 and 3), but four diverge (form 2); despite this the P1 Gln306 is remarkably fixed in position (side chain for Gln306 shown). d Structural alignment of all 74 S1 binding sites (stick) projected behind the S1 pocket (P1-Gln space filling and pocket surface is that of C4, provided for context). The P1 residue (Gln306) sidechain atoms are shown as semitransparent spheres (carbon - green, nitrogen - blue, oxygen - red). e Analogous all structure alignment of the S2 binding sites with space filling P2 (Leu305) and surface for the S2 pocket of C4. f Analogous all structure alignment of the S4 binding sites with space filling P4 (Ala303) and surface for the S4 pocket of C4.