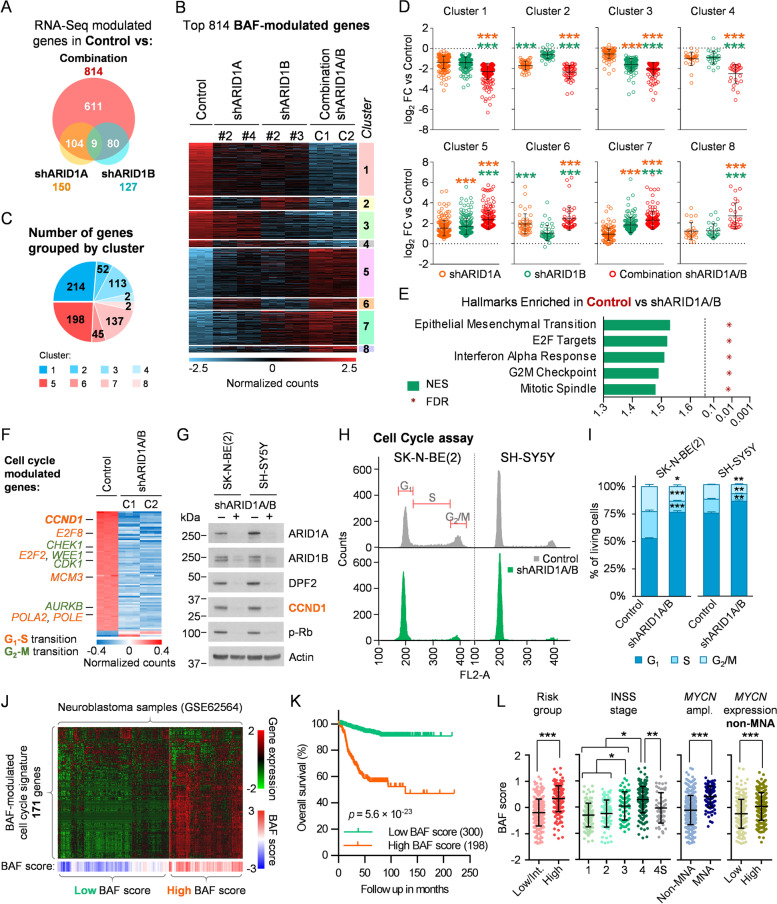
Fig. 2.
Co-depletion of ARID1A and ARID1B uncovers a wide transcriptome reprogramming involving cell cycle blockade. A Modulated genes in SK-N-BE(2) cells after single silencing of ARID1A or ARID1B with two different shRNAs for each gene (shARID1A, shARID1B) and two combinations of shRNA for both genes (Combination) analysed by RNA-Seq. Venn diagram shows the number of modulated genes in comparison with a non-silencing control for each of the three experimental conditions, using cutoff values of log2 FC (Fold Change) > 1.5 or < -1.5, and adjusted p-value < 0.001. B Heatmap representing relative RNA expression of genes modulated after BAF-disruption (Combination), using a cutoff of log2 FC > 1.5 or < -1.5, and adjusted p-value < 0.001. Genes were sorted by single inhibition behaviour-based clusters. C Pie chart representing the proportion of BAF-modulated genes included in each cluster. D Comparison of expression FC against control among experimental groups of BAF-modulated genes split in single inhibition behaviour-based clusters. Colour code indicates the condition against which the multiple comparison statistical tests are calculated. E Normalized Enrichment Score (NES) and False Discovery Rate (FDR) of the top 5 hallmarks of transcriptionally enriched genes in control compared with BAF-depleted cells (shARID1A/B), by Gene Set Enrichment Analysis. F Heatmap representing relative RNA expression of those genes included in E2F targets, G2-M checkpoint and mitotic spindle hallmarks from MSigDB, and modulated after BAF-disruption using a cutoff of log2 FC > 1 or < -1, and adjusted p-value < 0.001. G Cyclin D1 (CCND1) and phosphorylated Rb (p-Rb) western blot analysis of BAF-disrupted cells, at 96 h after transduction with shARID1A and shARID1B. H Cell cycle analysis of neuroblastoma cell lines co-transduced with shARID1A and shARID1B, or with control shRNA, analyzed by flow cytometry. I Quantification and comparison of the different cell cycle phase populations detected by flow cytometry. J RNA expression of a BAF-modulated the cell cycle transcriptional signature consisting of 171 genes in a gene expression dataset of 498 human neuroblastoma tumours (GSE62564). Patients were unbiasedly clustered into high and low BAF score groups, as described in Supplementary Materials and Methods. K Kaplan–Meier plots comparing the overall survival of high and low BAF score group of patients. L Comparison of BAF signature score according to risk groups, INSS (International Neuroblastoma Staging System) stages, MYCN amplification (MNA) and MYCN mRNA expression (above or below median) in non-MNA cases. * means p < 0.05; ** means p < 0.01; *** means p < 0.001