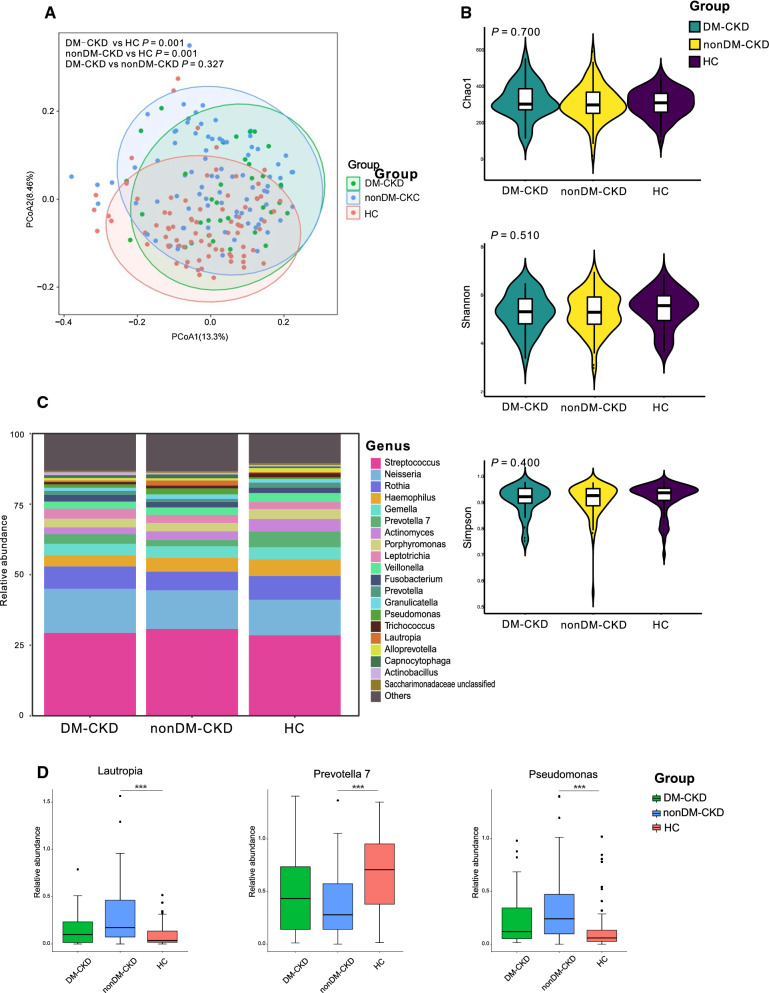
Fig. 4.
Association of diabetes in the salivary microbiome in CKD A. PCoA based on Bray–Curtis distances at ASV level showed different microbial compositions between groups of DM-CKD patients and HC (P < 0.05); between nonDM-CKD patients and HC(P < 0.05); between DM-CKD patients and nonDM-CKD patients (P > 0.05). Permutational multivariate analysis of variance (PERMANOVA) was performed for statistical comparisons of samples in the two cohorts. P-value was adjusted by the Benjamini and Hochberg false discovery rate (FDR). B. Bacterial richness and diversity measured by Chao1, Shannon, and Simpson were calculated at the microbial ASV level. Wilcoxon rank-sum test was performed and adjusted by Benjamini and Hochberg false discovery rate (FDR). C. Microbial profile at the genus level. The stacked plot demonstrated the top 15 most abundance bacterial genera in the groups of HC, DM-CKD, and nonDM-CKD. D. Bacterial genus that was differentially abundant among groups of HC, DM-CKD, and nonDM-CKD. Only the genus with above 1% is displayed. P-value was calculated using Wilcoxon rank-sum test and adjusted by Benjamini and Hochberg’s false discovery rate (FDR). *** indicate P < 0.001