Fig. 3.
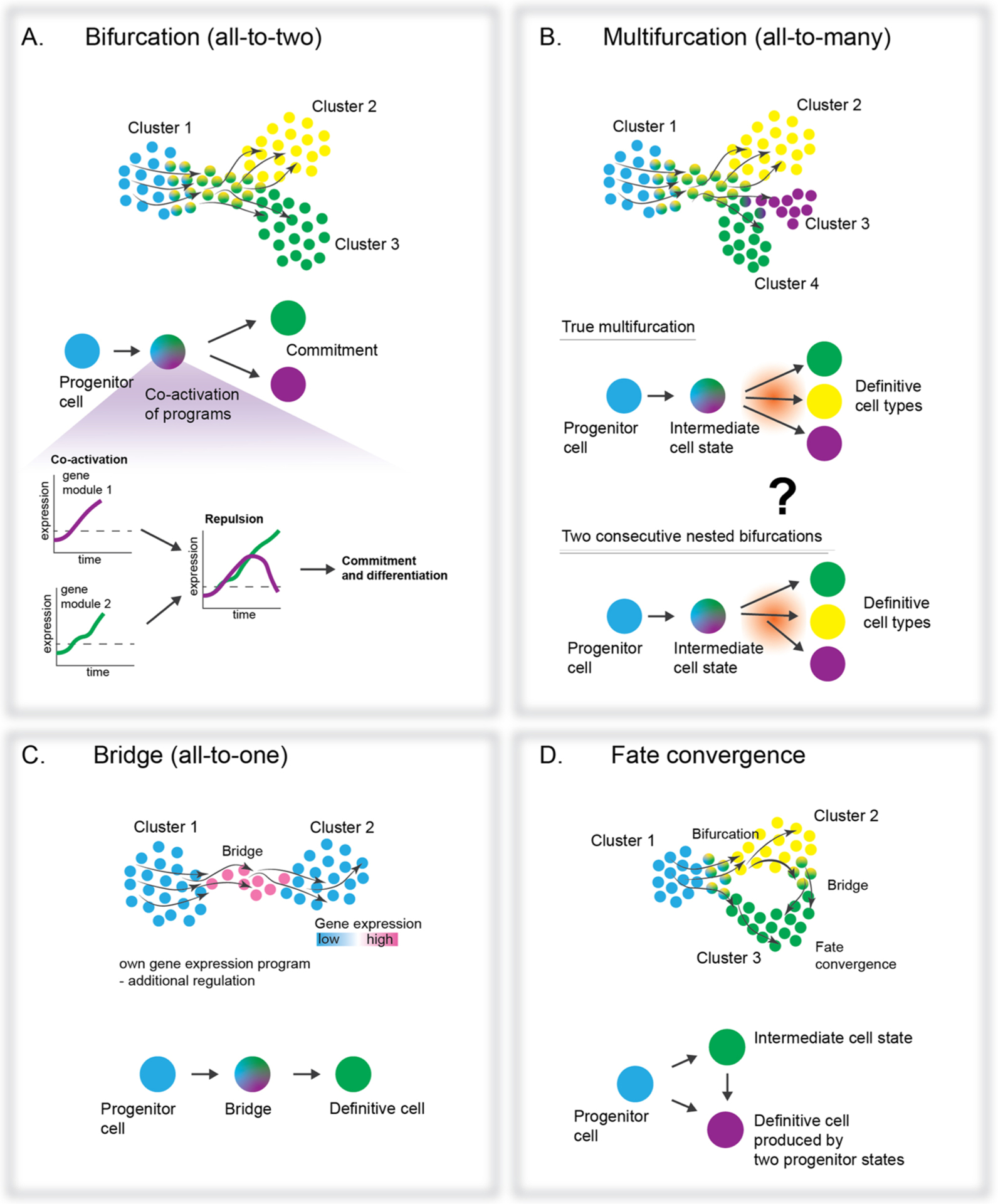
The diversity of transitions revealed on transcriptional landscapes. Single cell transcriptomics of cell populations, coupled with RNA velocity analysis, visualizes transcriptional dynamics as vector fields within a projected 2D transcriptional space, making it possible to assess features of the NCC fate decisions and resulting transitions. At least three motifs emerged as basic units representing transitions on the transcriptional landscape: bifurcation, multifurcation (still missing a reliable proof of existence), bridge, and fate convergence. A. Bifurcation occurs when a population of precursor cells decides between two distinct fates. The process features co-activation of fate-specific programs within each cell causing the development of micro-heterogeneity, and cross-repulsion of the competing programs. Eventually, one program wins, being immediately followed by fate segregation at the transcriptional level. B. Multifurcation is similar to a bifurcation with the number of potential fates being more than two. However, some uncertainty remains whether currently predicted multifurcations are in fact closely nested bifurcations (see question mark). C. Analysis of transient cell states have identified and transcriptionally characterized the intermediate cell states (bridges) that mediate the transition between progenitors and definitive cell populations. D. NCCs (and other progenitors in the body) often take different transcriptional routes to reach the same endpoint (definitive fate) resulting in convergence. These routes may feature differential susceptibility to extrinsic signaling events, which offers an additional regulatory level on clonal diversification.
