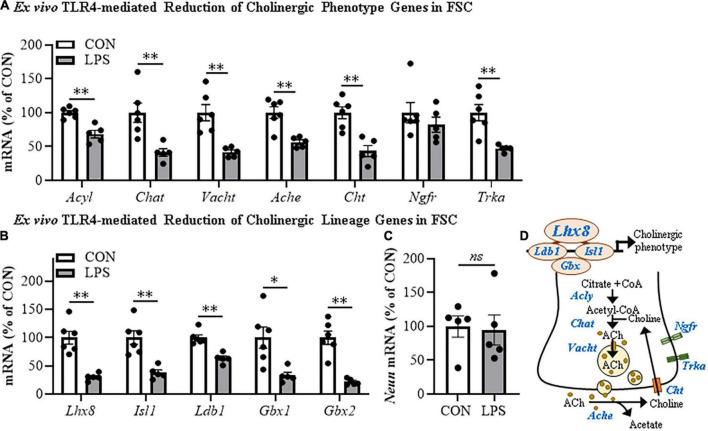
FIGURE 2.
Direct TLR4 activation with LPS reduces cholinergic neuron gene expression in the FSC model. Reverse transcription PCR (RTPCR) analysis revealed that direct application of LPS (100 ng/mL; 24 h) to basal FSC media decreased mRNA expression of (A) cholinergic phenotype (Acyl [31% (±6%)], Chat [59% (±6%)], Vacht [58% (±3%)], Ache [43% (±3%)], Cht [57% (±8%)], Trka [53% (±2%)]) and (B) lineage genes (Ldb1 [37% (±4%)], Isl1 [62% (±5%)], Lhx8 [70% (±3%)], Gbx1 [67% (±6%)], Gbx2 [78% (±2%)]) relative to vehicle-treated CONs. n = 5–6 wells/group. (C) RTPCR analysis revealed that direct application of LPS to FSC media did not affect mRNA expression of neuronal Neun mRNA relative to vehicle-treated CONs (n = 5 wells/group). (D) Simplified schematic depicting cholinergic phenotype and lineage genes in a basal forebrain cholinergic neuron. RTPCR analyses were run in duplicate. Data are presented as mean ± SEM. *p < 0.05, **p < 0.01, two-tailed t-tests. Ache, acetylcholinesterase; Acly, ATP citrate lyase; Chat, choline acetyltransferase; Cht, high-affinity choline transporter; Gbx, gastrulation brain homeobox; Isl1, insulin gene enhancer protein 1; Ldb1, LIM domain-binding protein 1; Lhx8, LIM/homeobox protein 8; Ngfr, nerve growth factor receptor; Trka, tropomyosin receptor kinase A; Vacht, vesicular acetylcholine transporter.