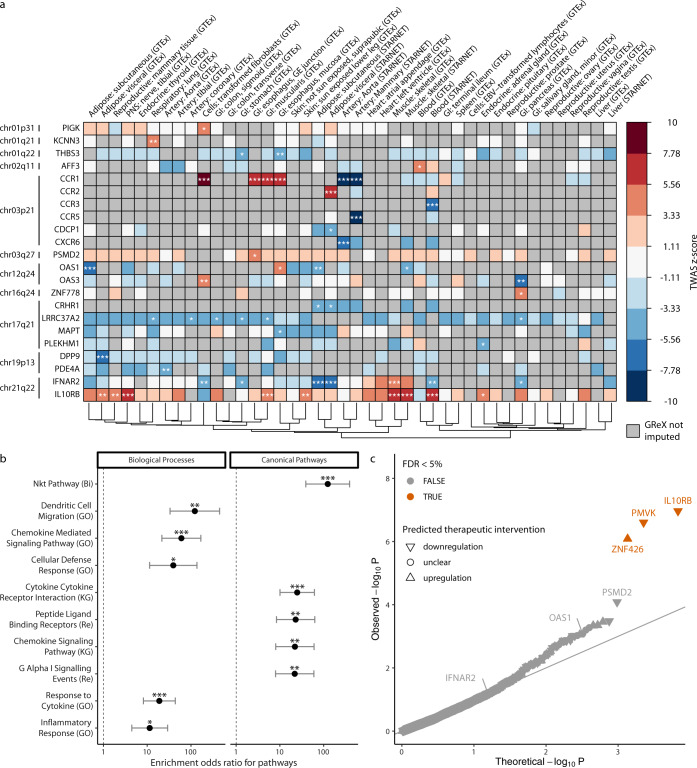
Fig. 2. Transcriptome-Wide Association Study (TWAS) for COVID-19-associated hospitalization (hospitalized COVID vs. the general population) identifies associated genes, pathways, and aids in identification of druggable gene targets.
a FDR-significant TWAS results for COVID-19 susceptibility across all tissues. Box color indicates gene-trait-tissue association z-scores. Gray squares represent genes whose genetically regulated gene expression (GReX) could not be imputed. ***, **, and * correspond to FDR-adjusted p-values of association equal or less than 0.001, 0.01, and 0.05 respectively. Dendrogram on the bottom edge is shown from Ward hierarchical clustering for tissues based on all GReX (not just FDR-significant results). Displayed results are limited to protein-coding genes; cytogenetic location (at band level resolution) is also provided on the left of each gene. b Enrichment of COVID-19 TWAS associated genes for biological processes and canonical pathways. Odds ratio with 95% confidence interval (CI) is plotted for the significant enrichments of TWAS gene-trait associations from all tissues. Pathways are ranked based on estimated enrichment odds ratio. Analysis is limited to protein-coding genes and excludes genes residing in the major histocompatibility complex (MHC) on chromosome 6. Enrichments that are FDR significant are annotated as follows: *, **, and *** for FDR-adjusted p ≤ 0.05, 0.01, and 0.001 respectively; Fisher’s exact test. c Prioritization of candidate gene targets to reverse TWAS gene-trait associations. p-value is estimated based on the joint statistic of two approaches () against the null. FDR-significant candidate genes are labeled orange. The direction of the predicted therapeutic intervention (upregulation or downregulation) is illustrated. IL10RB, PMVK, and ZNF426 are FDR-significant target genes (n = 4302 imputed genes with reliable shRNA signatures).