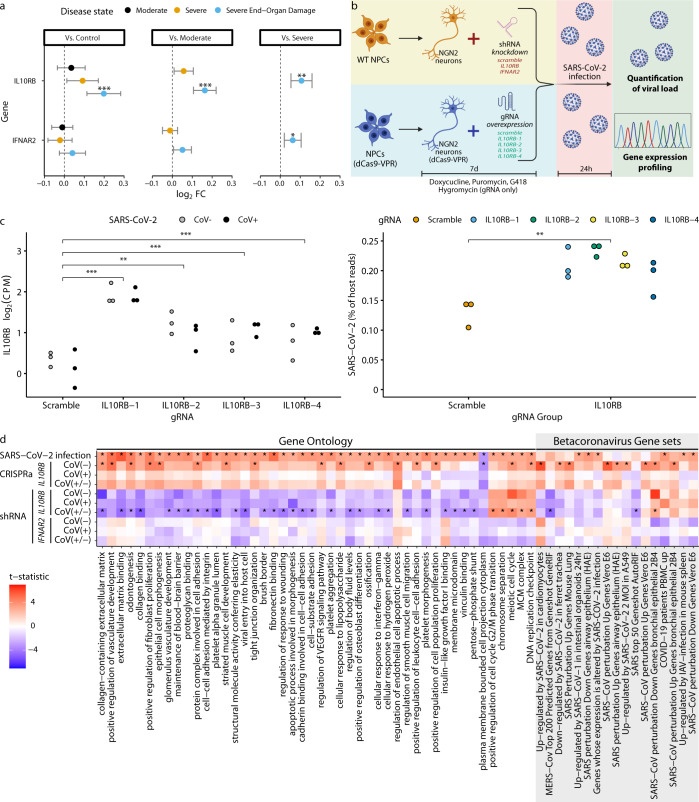
Fig. 4. Increased IL10RB expression is associated with worse COVID-19 outcomes in vivo and increased SARS-CoV-2 viral load in vitro.
a Increased IL10RB expression is associated with worse COVID-19 outcomes in vivo. *, **, and *** for FDR-adjusted p (FDR) ≤ 0.05, 0.01, and 0.001, respectively. Error bars show 95% CI. b In vitro experimental overview. c CRISPRa gRNAs (IL10RB-1, IL10RB-2, IL10RB-3, and IL10RB-4) were used to overexpress IL10RB in hiPSC-derived NGN2-glutamatergic neurons. ***, **, and * correspond to p-values from the linear model of equal or less than 0.001, 0.01, and 0.05, respectively. For the SARS-CoV-2 viral load (right panel) we performed pairwise comparison with unpaired t-test; ***, **, and * correspond to p values equal to, or less than, 0.001, 0.01, and 0.05, respectively. d Competitive gene set enrichment analysis in hiPSC-derived NGN2 glutamatergic neurons. Each row represents a different experimental condition and each column a different gene set; the top row shows the effect of SARS-CoV-2 infection, while the remaining rows show the effect of gene manipulation (e.g., IL10RB vs. nontargeting siRNA) within a specific group (e.g., CoV(−): uninfected cells). The left side of the plot (Gene ontology gene sets; white background) indicates enrichment for canonical pathways and biological processes that are significantly enriched (FDR < 0.05) in SARS-CoV-2 infection (top row), while the right side (Betacoronavirus Gene sets; gray background) illustrates enrichment for gene sets that correspond to betacoronavirus infections across different cell systems21 (only significant results are shown; FDR < 0.05).