Figure 1. LINC00839 is highly expressed in CRC and positively associated with poor outcomes in patients.
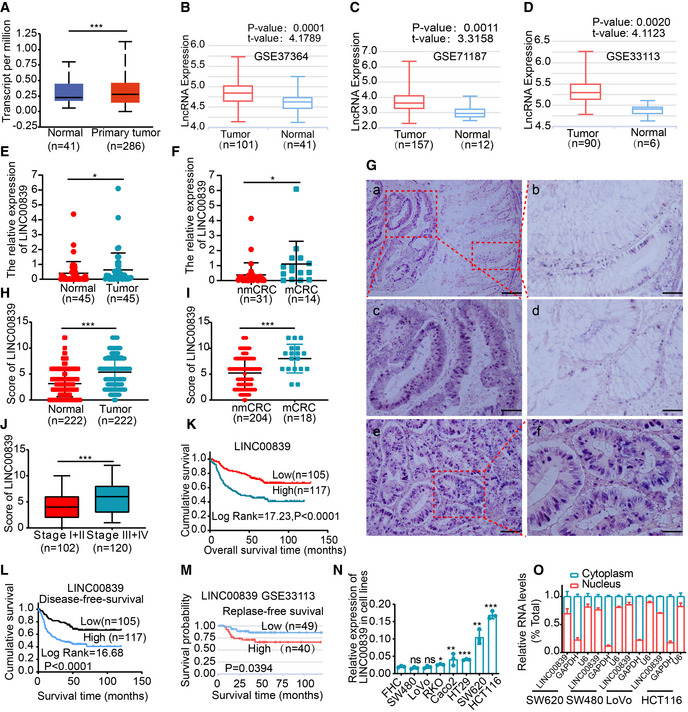
-
AThe expression of LINC00839 in CRC tissues and normal mucosa tissues in the UALCAN database.
- B–D
-
E, FThe expression levels of LINC00839 in tumor tissues and NATs from the CRC cohort (n = 45). nmCRC denotes CRC patients without metastases, and mCRC denotes CRC patients with metastases.
-
GRepresentative ISH staining of LINC00839 in paraffin‐embedded samples of CRC tumor tissues and NATs. (a) Expression of LINC00839 in tumor tissues and NATs. Scale bars, 100 μm. (b) Magnification of normal mucosal area and (c) magnification of the tumor area. Scale bars, 50 μm. (d) Weakly positive staining of LINC00839 in normal mucosa. Scale bars, 50 μm. (e) Strong positive staining of LINC00839 in tumors and (f) magnification of the local area. Scale bars, 100 μm.
-
HThe expression score of the tumors and NATs in the ISH cohort (n = 222).
-
IDifferential expression of LINC00839 in nonmetastatic CRC patients (nmCRC) and metastatic CRC patients (mCRC).
-
JExpression of LINC00839 in CRC patients with early‐stage (stage I and II) and advanced‐stage (stage III and IV) disease.
-
K, LKaplan–Meier analysis of overall survival and disease‐free survival rate of the ISH CRC cohort.
-
MKaplan–Meier analysis of the disease‐free survival rate of CRC patients in the LnCAR database (GSE33113).
-
NThe expression of LINC00839 in a colon mucosa epithelial cell line (FHC) and CRC cell lines.
-
OThe subcellular localization was determined by nucleocytoplasmic fractionation of several CRC cell lines.
Data information: In (A, B, C, D and J), the middle lines represent the median, and the upper and lower lines represent the upper and lower quartiles. In (E, F, N and O), the data are presented as the mean ± SD and were analyzed by Student's t‐test, n = 3 technical replicates. In (H and I), the data are shown as the mean ± SD and were analyzed by Student's t‐test. Across experiments, the P‐values are as follows: ns = not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001.
