Abstract
Ahnak, a large protein first identified as an inhibitor of TGF-β signaling in human neuroblastoma, was recently shown to promote TGF-β in some cancers. The TGF-β signaling pathway regulates cell growth, various biological functions, and cancer growth and metastasis. In this study, we used Ahnak knockout (KO) mice that underwent a 70% partial hepatectomy (PH) to investigate the function of Ahnak in TGF-β signaling during liver regeneration. At the indicated time points after PH, we analyzed the mRNA and protein expression of the TGF -β/Smad signaling pathway and cell cycle-related factors, evaluated the cell cycle through proliferating cell nuclear antigen (PCNA) immunostai-ning, analyzed the mitotic index by hematoxylin and eosin staining. We also measured the ratio of liver tissue weight to body weight. Activation of TGF-β signaling was confirmed by analyzing the levels of phospho-Smad 2 and 3 in the liver at the indicated time points after PH and was lower in Ahnak KO mice than in WT mice. The expression levels of cyclin B1, D1, and E1; proteins in the Rb/E2F transcriptional pathway, which regulates the cell cycle; and the numbers of PCNA-positive cells were increased in Ahnak KO mice and showed tendencies opposite that of TGF-β expression. During postoperative regeneration, the liver weight to body weight ratio tended to increase faster in Ahnak KO mice. However, 7 days after PH, both groups of mice showed similar rates of regeneration, following which their active regeneration stopped. Analysis of hepatocytes undergoing mitosis showed that there were more mitotic cells in Ahnak KO mice, consistent with the weight ratio. Our findings suggest that Ahnak enhances TGF-β signaling during postoperative liver regeneration, resulting in cell cycle disruption; this highlights a novel role of Ahnak in liver regeneration. These results provide new insight into liver regeneration and potential treatment targets for liver diseases that require surgical treatment.
Keywords: 70% partial hepatectomy, Ahnak KO mice, Hepatocyte proliferation, Liver regeneration, TGF-β/Smad signaling pathway
INTRODUCTION
Ahnak, a large, 700 kDa protein that was first identified in human neuroblastoma and skin epithelial cells, is a well-known inhibitor of TGF-β signaling (1). Due to its large size, Ahnak can bind multiple proteins and mediate signaling (2, 3). Ahnak has distinct roles in different cancers. For instance, Ahnak was downregulated in neuroblastoma but upregulated in glioma, mesothelioma, fibrosarcoma, and prostate cancer (1, 4). Ahnak functions as a tumor suppressor in lung tumors, breast tumors, and hepatocellular carcinoma (5, 6). Activation of TGF-β facilitates hepatocellular infiltration and metastasis. Ahnak functions as a tumor suppressor by promoting TGF-β/Smad3 signaling to arrest cell growth (7, 8). These results suggest that Ahnak regulates the growth of cells, including tumor cells, and that this activity is related to TGF-β. TGF-β has a crucial role in inhibiting hepatocellular proliferation during the regeneration of acutely damaged livers (8, 9). Immediately after acute damage, such as partial hepatectomy, the liver actively regenerates, without necrosis or inflammation. Within 7 days after surgery, the liver returns to its pre-surgery size to maintain biological homeostasis (10). TGF-β induces a reduction in DNA synthesis during liver regeneration, which ultimately stops regeneration. TGF-β1 is the most important form of TGF-β in this process (11, 12). Although it is known that TGF-β1 is involved in liver regeneration, the termination process is still poorly understood, thus, it is expected that Ahnak participates in cell growth regulation, especially cessation of liver regeneration (8, 10). Many studies have used genetic methods to explore the process of liver regeneration (13). However, the role of Ahnak has not been explored. Therefore, we used Ahnak KO mice that underwent a 70% PH to investigate the role of Ahnak in liver regeneration (14, 15). TGF-β expression and the liver to body weight ratio were analyzed, cell proliferation and mitosis were evaluated by proliferating cell nuclear antigen (PCNA) immunostaining and hematoxylin and eosin (H&E) staining, and the expression of cell cycle-related cyclins and transcription factors were evaluated. The findings of this study are expected to improve our understanding of the roles of Ahnak in cell growth and the molecular biology of liver regeneration.
Furthermore, in the liver regeneration process induced by acute injury, the function of cytokines such as TNF-α and IL-6 that promote regeneration and growth factors such as HGF, EGF, TGF, insulin, and glucagon are relatively clear, but the mechanisms of the termination process in liver regeneration remain unclear (10, 16). Therefore, an investigation of Ahnak that regulates TGF-β is required to understand the regeneration termination process that is not explained by both TGF-β and Activin, which are well-known antiproliferative factors (16).
RESULTS
Ahnak depletion influences the TGF-β/Smad signaling pathway in liver regeneration after 70% partial hepatectomy
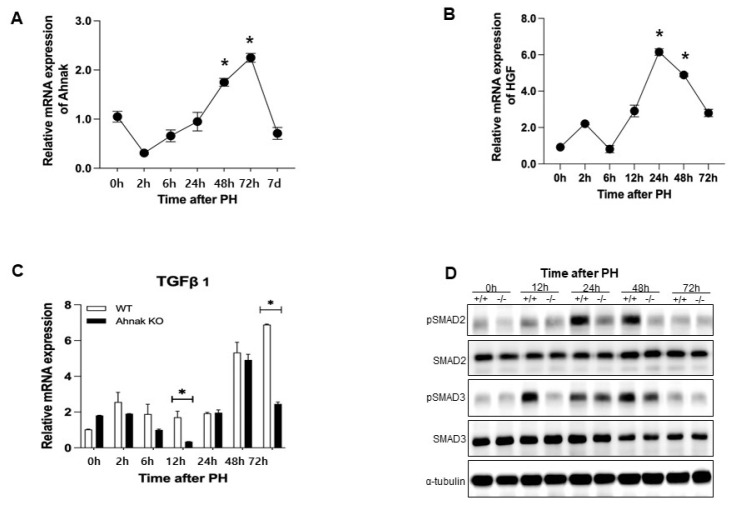
To investigate the impact of Ahnak on liver regeneration, we performed a 70% PH in WT mice and assessed Ahnak mRNA expression by RT-PCR at the indicated time points after PH. Immediately after surgery, Ahnak expression was lowest and was lower than preoperative (Fig. 1A). Expression was increased from 24 h after PH and then gradually decreased from 72 h after PH (Fig. 1A). The Ahnak expression pattern was the same as the patterns of such molecule which HGF (hepatocellular growth factor) known to regulate liver regeneration, which change in a time-dependent manner after PH (Fig. 1B) (17).
Fig. 1.
Ahnak depletion influences the TGF-β1/Smad signaling pathway during liver regeneration after partial hepatectomy (PH). (A) Ahnak mRNA expression in wild type (WT) mice at different time points after PH (n = 3). (B) HGF mRNA expression in wild-type (WT) mice at different time points after PH (n = 3). (C) TGF-β1 mRNA expression in Ahnak knockout (KO) and WT mice at the indicated time points after PH (n = 3). (D) Representative western blots of SMAD2, phospho-SMAD2, SMAD3, and phospho-SMAD3 at the indicated time points. *P < 0.05; Error bars represent the mean ± SD.
To determine the role of Ahnak in liver regeneration, we also performed a 70% PH in Ahnak KO mice and analyzed TGF-b mRNA expression at the indicated time points after PH. TGF-β is a mitosis-related inhibitor of cell growth that is involved in the termination of liver regeneration (8). It is known that Ahnak-mediated heighten of TGF-β signaling induces negative regulation of cell growth (7). During liver regeneration, TGF-b expression was significantly lower in Ahnak KO mice than in WT mice (Fig. 1C).
TGF-β functions by binding to the TGF-β type II receptor, which phosphorylates the type I receptor. Activated type I receptor phosphorylates the R-Smads (receptor-regulated Smads) such as Smad2 and Smad3. Phosphorylated Smad2 and Smad3 combine with Smad4 (a co-Smad), which then moves into the nucleus where it regulates the transcription of target genes (18). Therefore, Smad expression is an indicator of TGF-β activation; therefore, we examined Smad2 and Smad3 in both WT and Ahnak KO mice after PH. During liver regeneration, Smad activation was lower in Ahnak KO mice than in WT mice, and Smad3 was downregulated earlier than Smad2 in the Ahnak KO mice. (Fig. 1D). There was a notable difference in Smad activation over the first 12-48 h after PH between Ahnak KO mice and WT mice. These results suggest that Ahnak regulates activation of the TGF-β/Smad signaling pathway and may influence cell growth during liver regeneration after PH.
Ahnak knockout promotes cell cycle progression and hepatocyte proliferation during liver regeneration after PH in mice
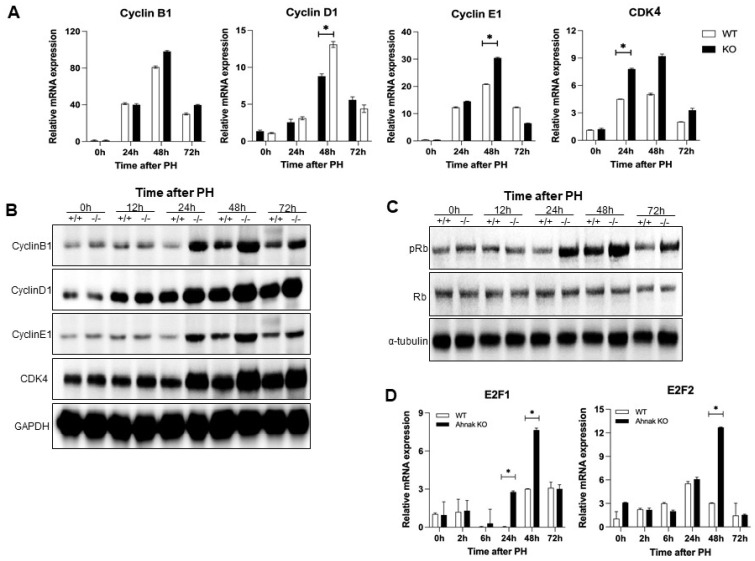
To confirm the effect of Ahnak knockout on the cell cycle during liver regeneration after PH, we examined the cell cycle-associated cyclins D1, E1, and B1 and Rb/E2F pathway in both WT and Ahnak KO mice (19). Cyclins are indicators of the regenerative activity of hepatocytes. Cyclin D1 plays a prominent role in proliferation and growth and is the most reliable marker for G1 progression in hepatocytes. CDK4, a cyclin-dependent kinase and key regulator of the cell cycle, forms a complex with cyclin D1. Cyclin E1 is associated with S phase progression, and cyclin B1 is associated with M phase (9). Cyclin D1 and CDK4 levels at 24-72 h after PH were higher in Ahnak KO mice than in WT mice. Cyclin E1 and cyclin B1 expression patterns were similar to that of cyclin D1 (Fig. 2A, B) (9, 20). As shown in Fig. 2A, B, although the time-dependent changes in mRNA and protein expression levels during liver regeneration do not exactly overlap, the trends are similar.
Fig. 2.
Knockout of Ahnak upregulates the expression of cell cycle-related genes in the liver after PH. (A) Cyclin and cell cycle-related mRNA expression levels in the liver at the indicated time points after PH. (B) Representative western blot of cyclins B1, D1, and E1 and CDK4 protein levels in the liver at the indicated time points after PH. (C) Representative western blot of Rb and phospho-Rb levels in the liver at the indicated time points after PH. (D) E2F1 and E2F2 mRNA expression levels in the liver at the indicated time points after PH (n = 3). *P < 0.05; Error bars represent the mean ± SD.
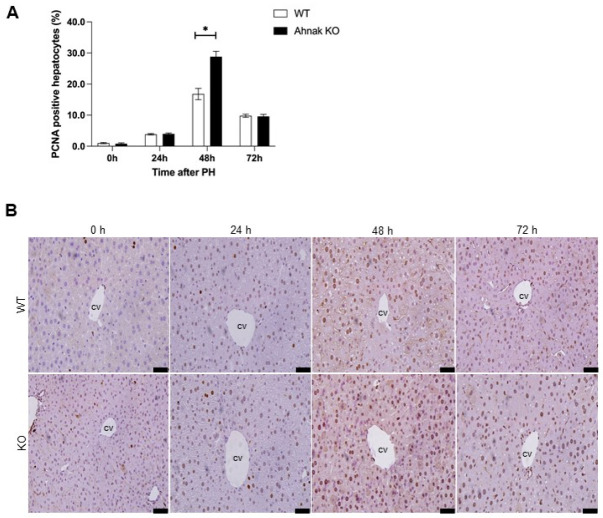
To understand the influence of Ahnak on cell cycle progression, we next evaluated Rb and E2F expression. E2Fs are transcription factors that regulate genes involved in DNA replication and cell cycle progression along with phosphorylated Rb (19, 21). Activation of Rb, as evaluated by the levels of the phosphorylated form, was higher in Ahnak KO mice than in WT mice over the first 24-72 h after PH (Fig. 2C). The mRNA expression levels of E2F1 and E2F2 were significantly higher in Ahnak KO mice than in WT mice at 48 h after PH (Fig. 2D). We then conducted an additional experiment to analyze the effect of Ahnak on cell proliferation using immunostaining of PCNA as a marker of cell cycle progression (Fig. 3B) (22). The rate of PCNA-positive hepatocytes was significantly higher in Ahnak KO mice than in WT mice at 48 h after PH (Fig. 3A). These results suggest that Ahnak modulates the cell cycle and cell proliferation during liver regeneration after PH.
Fig. 3.
Knockout of Ahnak accelerates hepatocyte proliferation in the liver after PH. (A) Quantification of proliferating cell nuclear antigen (PCNA) staining in hepatocyte nuclei at the indicated time points after PH (n = 3). (B) Representative immunohistochemical images showing PCNA staining at the indicated time points after PH. *P < 0.05; Error bars represent the mean ± SD. Scale bar = 50 μm.
Ahnak knockout upregulates mitosis and accelerates liver regeneration
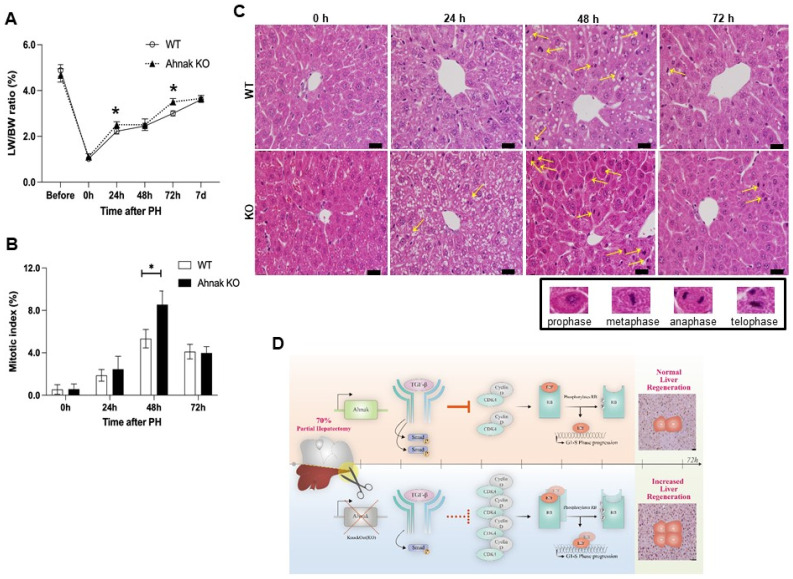
To evaluate hepatocyte mitosis during liver regeneration, we analyzed the hepatocyte mitotic index using H&E staining (Fig. 4C). The number of hepatocytes undergoing mitosis was dramatically increased in Ahnak KO mice compared to the number in WT mice at 48 h after PH (Fig. 4B). The mitotic index was calculated by counting the number of nuclei in each phase of mitosis (prophase, metaphase, anaphase, and telophase) (see yellow arrow in Fig. 4C).
Fig. 4.
Ahnak knockout (KO) mice show increased mitosis in the liver after PH. (A) Liver to body weight ratios at the indicated time points after PH in WT and Ahnak KO mice (n = 3, each). (B) Quantification of hepatocyte nuclei undergoing mitosis at the indicated time points after PH (n = 3). (C) Representative hematoxylin-eosin staining images and nuclei during mitosis (yellow arrow) in the liver at the indicated time points after PH. (D) Graphical summary of Ahnak expression in liver regeneration after PH. *P < 0.05; Error bars represent the mean ± SD; Scale bar = 10 μm.
Finally, we evaluated liver regeneration by calculating the ratio of liver weight to body weight for 7 days after PH. Although alterations in gene expression persist for up to 14 days after PH, hepatic parenchymal growth is most active for the first week after PH (16). The ratio was significantly higher in Ahnak KO mice than in WT mice at 24 and 48 h after PH (Fig. 4A). The ratio reached 83% of the pre-surgery liver mass within 7 days after PH in both Ahnak KO and WT mice.
DISCUSSION
The liver is the only organ with regenerative ability, and the liver-to-body weight ratio is maintained at the required level (100%) for homeostasis (17, 23).
To study this crucial regenerative ability, a 70% partial hepatectomy (PH) has been widely used as a model of acute liver damage. This model has been used to reveal the extracellular and intracellular signaling mechanisms involved in the process of restoring the liver to its pre-surgery size and weight (16, 23). Many studies have revealed several pathways related to liver regeneration. However, the detailed mechanisms have not yet been determined. In particular, the mechanism of regeneration termination is not well understood (10, 23). Therefore, we focused on Ahnak, which is associated with the TGF-β signaling involved in the termination phase of liver regeneration. First, we analyzed the Ahnak mRNA expression pattern after PH in WT mice, which showed that expression gradually increased and then reversibly decreased to pre-surgery levels, and this pattern was similar to those of genes that regulate liver regeneration, suggesting that Ahnak may regulate liver regeneration (13, 24-26). Next, we analyzed activation of the TGF-β/Smad signaling pathway by measuring the activation of SMAD2 and SMAD3 (27, 28). Smad activation was significantly reduced in Ahnak KO mice when compared to WT mice. During recovery after acute liver damage, TGF-β inhibits the growth factor-induced signals of cyclin D1 and E1, which are activators of G1 phase (29). As expected, our results showed that a reduction in TGF-β1 signaling influences these cell cycle-related genes in Ahnak KO mice. In addition, the expression of transcription factor CDK4, its complex partner cyclin D1, and cyclin E1, which promote cell cycle progression, were also increased in Ahnak KO mice. The levels of Rb protein, which stimulates the transcription factor E2F in late G1 phase and regulates S phase of the cell cycle, were also higher in Ahnak KO mice than in WT mice (30). Expression of cyclin B1, which promotes M phase, was also higher in Ahnak KO mice. The increase in the PCNA-positive rate in Ahnak KO mice after PH indicated active cell cycle progression. These results show that the loss of Ahnak promotes cell proliferation. Finally, we observed that the ratios of liver to body weight and hepatocytes in mitosis were higher at specific time points after PH in Ahnak KO mice than in WT mice, suggesting that the molecular mechanism of hepatocyte proliferation was activated in Ahnak KO mice. Although the liver-to-body weight ratios in Ahnak KO and WT mice were similar at 48h after PH (Fig. 4A), cell proliferation was more active in Ahnak KO mice over the first 24-72 h after surgery (Fig. 3A and 4B). DNA replication and hepatocyte hyperplasia achieve liver mass restoration 24-96 h after PH in mice (17, 31). Although some inconsistency between the experimental groups was observed, the degree of the regeneration rate was evaluated based on the trend in the regenerative process.
Our findings support those of a previous study showing that Ahnak promotes the TGF-β/Smad signaling pathway, which inhibits cell cycle progression. One fascinating result is that Ahnak activates TGF-β signaling during liver regeneration after PH (Fig. 4D). This is because the results are contrary to the effects found in previous studies of hepatocellular carcinoma (6).
In a previous study using hepatocyte-specific TGF-β type II receptor gene KO mice showed that an increase in regenerating hepatocyte levels via the acceleration of G1-S phase cell cycle transition was observed, but the liver repair rate on termination was normal (32, 33). We confirmed that the liver repair rates in Ahnak KO and WT mice were similar and that liver regeneration was nearly complete at 7 days after PH. Contrary to expectations, this finding indicates that Ahnak is not the most critical gene in stopping liver regeneration. Our research offers an additional information to understand liver regeneration. Therefore, continued research on this termination pathway in liver regeneration is necessary.
We found that systemic deletion of Ahnak affects liver regeneration and suggest that Ahnak may be useful for monitoring the recovery progress of patients who undergo liver resection. Considering the impact of this study, we are planning to examine the role of Ahnak after surgical resection for hepatic tumors.
MATERIALS AND METHODS
Animals
Ahnak KO mice were created by disrupting exon 5 of the Ahnak gene, as described by Lee (34). Genotyping was performed using genomic DNA isolated from the tail according to the methods described by Lee (34). Ahnak KO mice were obtained by crossing heterozygous breeders. Mice were maintained under a 12 h light-dark cycle and were supplied free access to water and a regular chow diet in a specific pathogen free facility.
Partial hepatectomy
Male 8-10-week-old mice were subjected to 70% PH under isoflurane inhalation anesthesia according to a published protocol (14, 15).
The left lateral lobe and median lobe of the liver with the gall bladder were ligated and removed. The gall bladder was removed during surgery to avoid damaging it. For post-oper-ative care, all animals were administered ketoprofen (5 mg/kg intraperitoneally; Daehan, Korea) to control pain. Mice were sacrificed at the indicated time points. Remnant liver was measured by weight, fixed in 4% paraformaldehyde, and snap frozen in liquid nitrogen immediately after extraction. Animal experiments were performed in accordance with the “Guide for Animal Experiments” of the Korean Academy of Medical Sciences and were approved by the Institutional Animal Care and Use Committee of Seoul National University, Seoul, Korea (IACUC approval no. SNU-160211).
Histology and immunohistochemistry
Liver tissues were fixed overnight in 4% paraformaldehyde, embedded in paraffin, and used for H&E staining and immuno-staining with antibodies against PCNA. The immunostaining was developed with diaminobenzidine. To quantify hepatocyte proliferation after immunostaining, 10 fields per slide were randomly chosen under the microscope to count the PCNA-positive hepatocytes and calculate the percentage of PCNA-positive hepatocytes among the total hepatocytes in each field.
Mitotic hepatocytes were counted in 10 fields per slide of H&E-stained hepatocytes. Following the mitotic figures defined by Baak, mitotic cells were counted based on nuclear morphology under a microscope. The mitotic index was defined as the mean number of mitotic cells in 10 fields (35).
Western blotting
Protein lysates prepared in RIPA buffer containing 0.5 mM phenyl methane sulfonyl fluoride, 4 μg/ml leupeptin, 4 μg/ml aprotinin, and 4 μg/ml pepstatin, were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred to polyvinylidene fluoride membranes. The membranes were incubated overnight with primary antibodies against the following proteins: pSMAD2, SMAD2, pSMAD3, SMAD3, Cyclin B1, Cyclin D1, Cyclin E1, CDK4, pRb, Rb, GAPDH, and α-tubulin. The membranes were then incubated with a goat-anti-rabbit-HRP or goat-anti-mouse-HRP secondary antibody for 1 h. Antibody binding was visualized using the Pierce TM ECL western blotting detection system (GE Co.).
qRT-PCR
Total RNA was isolated from the liver using TRIzol (Ambion) reagent. Real-time RT-PCR analysis of the isolated mRNA was performed in a two-step reaction. The first strand complementary DNA was synthesized with the Acculower RT reverse transcription kit (Bioneer), and the second step was performed on a 7500 Real Time PCR System (Applied Biosystems) using SYBR green (BIO-94020; Bioline) and specific primers for each of the target genes. Every assay included the 36B4 gene as an endogenous reference. Results were analyzed with Student’s unpaired t-test. Gene expression was calculated using the 2−DDCT method.
Statistical analysis
Statistical analysis was performed using GraphPad Prism 4 (GraphPad Software, http://www.graphpad.com). Data are presented as mean ± standard error of the mean (SEM). Statistical significance among more than two groups was assessed using Student’s t test. A P value less than 0.05 was considered statistically significant.
ACKNOWLEDGEMENTS
This research was supported by Korea Mouse Phenotyping Project (2013M3A9D5072550) of the National Research Foundation (NRF) funded by the Ministry of Science and ICT (2012M3A 9D1054622) and partially supported by the Brain Korea 21 Plus Program and the Research Institute for Veterinary Science of Seoul National University.
Footnotes
CONFLICTS OF INTEREST
The authors have no conflicting interests.
REFERENCES
- 1.Shtivelman E, Cohen FE, Bishop JM. A human gene (AHNAK) encoding an unusually large protein with a 1.2-microns polyionic rod structure. Proc Natl Acad Sci U S A. 1992;89:5472–5476. doi: 10.1073/pnas.89.12.5472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Han H, Kursula P. Periaxin and AHNAK nucleoprotein 2 form intertwined homodimers through domain swapping. J Biol Chem. 2014;289:14121–14131. doi: 10.1074/jbc.M114.554816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dempsey BR, Rezvanpour A, Lee TW, Barber KR, Junop MS, Shaw GS. Structure of an asymmetric ternary protein complex provides insight for membrane interaction. Structure. 2012;20:1737–1745. doi: 10.1016/j.str.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 4.Shankar J, Messenberg A, Chan J, Underhill TM, Foster LJ, Nabi IR. Pseudopodial actin dynamics control epithelial-mesenchymal transition in metastatic cancer cells. Cancer Res. 2010;70:3780–3790. doi: 10.1158/0008-5472.CAN-09-4439. [DOI] [PubMed] [Google Scholar]
- 5.Park JW, Kim IY, Choi JW, et al. AHNAK loss in mice promotes type II pneumocyte hyperplasia and lung tumor development. Mol Cancer Res. 2018;16:1287–1298. doi: 10.1158/1541-7786.MCR-17-0726. [DOI] [PubMed] [Google Scholar]
- 6.Peng R, Zhang PF, Yang X, et al. Overexpression of RNF38 facilitates TGF-beta signaling by Ubiquitinating and degrading AHNAK in hepatocellular carcinoma. J Exp Clin Cancer Res. 2019;38:113. doi: 10.1186/s13046-019-1113-3.2bdf0fd58bac47ceb3d853a69ef950ff [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee IH, Sohn M, Lim HJ, et al. Ahnak functions as a tumor suppressor via modulation of TGFbeta/Smad signaling pathway. Oncogene. 2014;33:4675–4684. doi: 10.1038/onc.2014.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Michalopoulos GK. Liver regeneration after partial hepatectomy: critical analysis of mechanistic dilemmas. Am J Pathol. 2010;176:2–13. doi: 10.2353/ajpath.2010.090675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fausto N. Liver regeneration. J Hepatol. 2000;32:19–31. doi: 10.1016/S0168-8278(00)80412-2. [DOI] [PubMed] [Google Scholar]
- 10.Taub R. Liver regeneration: from myth to mechanism. Nat Rev Mol Cell Biol. 2004;5:836–847. doi: 10.1038/nrm1489. [DOI] [PubMed] [Google Scholar]
- 11.Böttinger EP, Factor VM, Tsang ML, et al. The recombinant proregion of transforming growth factor beta1 (latency-associated peptide) inhibits active transforming growth factor beta1 in transgenic mice. Proc Natl Acad Sci U S A. 1996;93:5877–5882. doi: 10.1073/pnas.93.12.5877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jakowlew SB, Mead JE, Danielpour D, Wu J, Roberts AB, Fausto N. Transforming growth factor-beta (TGF-beta) isoforms in rat liver regeneration: messenger RNA expression and activation of latent TGF-beta. Cell Regul. 1991;2:535–548. doi: 10.1091/mbc.2.7.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fausto N, Campbell JS, Riehle KJ. Liver regeneration. Hepatology. 2006;43:S45–53. doi: 10.1002/hep.20969. [DOI] [PubMed] [Google Scholar]
- 14.Higgins GM. Experimental pathology of the liver. I. Restoration of the liver of the white rat following partial surgical removal. Arch Pathol. 1931;12:186–202. [Google Scholar]
- 15.Mitchell C, Willenbring H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat Protoc. 2008;3:1167–1170. doi: 10.1038/nprot.2008.80. [DOI] [PubMed] [Google Scholar]
- 16.Michalopoulos GK. Hepatostat: liver regeneration and normal liver tissue maintenance. Hepatology. 2017;65:1384–1392. doi: 10.1002/hep.28988. [DOI] [PubMed] [Google Scholar]
- 17.Michalopoulos GK. Liver regeneration. J Cell Physiol. 2007;213:286–300. doi: 10.1002/jcp.21172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kokura K, Kim H, Shinagawa T, Khan MM, Nomura T, Ishii S. The Ski-binding protein C184M negatively regulates tumor growth factor-beta signaling by sequestering the Smad proteins in the cytoplasm. J Biol Chem. 2003;278:20133–20139. doi: 10.1074/jbc.M210855200. [DOI] [PubMed] [Google Scholar]
- 19.Kitamura T, Watanabe S, Sato N. Liver regeneration, liver cancers and cyclins. J Gastroenterol Hepatol. 1998;13:S96–S99. doi: 10.1111/jgh.1998.13.s1.96. [DOI] [PubMed] [Google Scholar]
- 20.LK M. Distinct proliferative and transcriptional effects of the D-type cyclins in vivo. Cell Cycle. 2008;7:2215–2224. doi: 10.4161/cc.7.14.6274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu HX, Fang Y, Hu Y, Gonzalez FJ, Fang J, Wan YJ. PPARbeta regulates liver regeneration by modulating Akt and E2f signaling. PLoS One. 2013;8:e65644. doi: 10.1371/journal.pone.0065644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Assy N, Gong Y, Zhang M, Pettigrew NM, Pashniak D, Minuk GY. Use of proliferating cell nuclear antigen as a marker of liver regeneration after partial hepatectomy in rats. J Lab Clin Med. 1998;131:251–256. doi: 10.1016/S0022-2143(98)90097-X. [DOI] [PubMed] [Google Scholar]
- 23.Michalopoulos GK, Bhushan B. Liver regeneration: biological and pathological mechanisms and implications. Nat Rev Gastroenterol Hepatol. 2021;18:40–55. doi: 10.1038/s41575-020-0342-4. [DOI] [PubMed] [Google Scholar]
- 24.Webber EM, Bruix J, Pierce RH, Fausto N. Tumor necrosis factor primes hepatocytes for DNA replication in the rat. Hepatology. 1998;28:1226–1234. doi: 10.1002/hep.510280509. [DOI] [PubMed] [Google Scholar]
- 25.Albrecht JH, Poon RY, Ahonen CL, Rieland BM, Deng C, Crary GS. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene. 1998;16:2141–2150. doi: 10.1038/sj.onc.1201728. [DOI] [PubMed] [Google Scholar]
- 26.Goggin MM, Nelsen CJ, Kimball SR, Jefferson LS, Morley SJ, Albrecht JH. Rapamycin-sensitive induction of eukaryotic initiation factor 4F in regenerating mouse liver. Hepatology. 2004;40:537–544. doi: 10.1002/hep.20338. [DOI] [PubMed] [Google Scholar]
- 27.Kogure K, Zhang YQ, Maeshima A, Suzuki K, Kuwano H, Kojima I. The role of activin and transforming growth factor-beta in the regulation of organ mass in the rat liver. Hepatology. 2000;31:916–921. doi: 10.1053/he.2000.6100. [DOI] [PubMed] [Google Scholar]
- 28.Wrighton KH, Lin X, Feng XH. Phospho-control of TGF-beta superfamily signaling. Cell Res. 2009;19:8–20. doi: 10.1038/cr.2008.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moriuchi A, Hirono S, Ido A, et al. Additive and inhibitory effects of simultaneous treatment with growth factors on DNA synthesis through MAPK pathway and G1 cyclins in rat hepatocytes. Biochem Biophys Res Commun. 2001;280:368–373. doi: 10.1006/bbrc.2000.4063. [DOI] [PubMed] [Google Scholar]
- 30.Nevins JR. The Rb/E2F pathway and cancer. Hum Mol Genet. 2001;10:699–703. doi: 10.1093/hmg/10.7.699. [DOI] [PubMed] [Google Scholar]
- 31.Marongiu F, Marongiu M, Contini A, et al. Hyperplasia vs hypertrophy in tissue regeneration after extensive liver resection. World J Gastroenterol. 2017;23:1764–1770. doi: 10.3748/wjg.v23.i10.1764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Oe S, Lemmer ER, Conner EA, et al. Intact signaling by transforming growth factor beta is not required for termination of liver regeneration in mice. Hepatology. 2004;40:1098–1105. doi: 10.1002/hep.20426. [DOI] [PubMed] [Google Scholar]
- 33.Romero-Gallo J, Sozmen EG, Chytil A, et al. Inactivation of TGF-beta signaling in hepatocytes results in an increased proliferative response after partial hepatectomy. Oncogene. 2005;24:3028–3041. doi: 10.1038/sj.onc.1208475. [DOI] [PubMed] [Google Scholar]
- 34.Lee IH, Lim HJ, Yoon S, et al. Ahnak protein activates protein kinase C (PKC) through dissociation of the PKC-protein phosphatase 2A complex. J Biol Chem. 2008;283:6312–6320. doi: 10.1074/jbc.M706878200. [DOI] [PubMed] [Google Scholar]
- 35.Baak JP. Mitosis counting in tumors. Hum Pathol. 1990;21:683–685. doi: 10.1016/0046-8177(90)90026-2. [DOI] [PubMed] [Google Scholar]