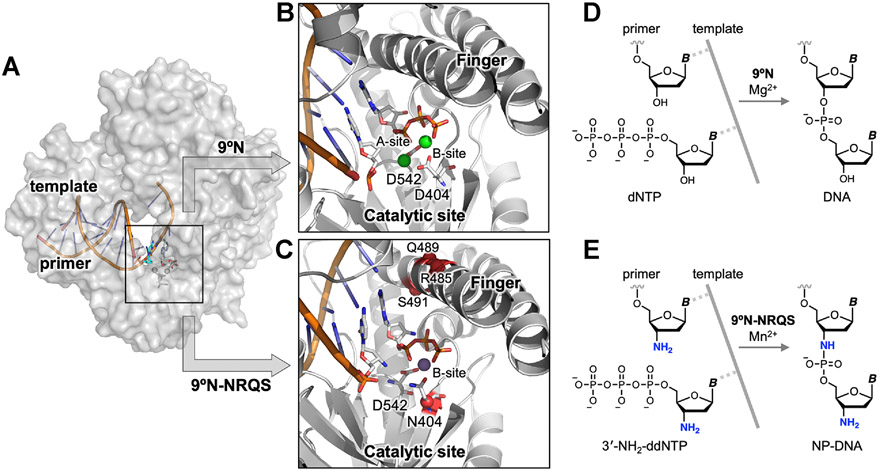
Figure 1.
Structure-guided redesign of 9°N DNA polymerase to catalyze the formation of internucleotidyl N-P bonds. (A) Model of 9°N bound to a primer/template complex. (B,C) Insets show the positioning of the primer and the incoming nucleoside triphosphate within (B) the WT 9°N (PDB accession codes: 5OMV and 5OMQ) and (C) its quadruple-mutant variant 9°N-NRQS (Rosetta based model). Sites of substitutions in 9°N-NRQS are highlighted by red spheres. Active site metal ions are shown: (B) Mg2+ in green and (C) Mn2+ in purple. Residues are drawn as sticks with color by atom (carbon, white; nitrogen, blue; oxygen, red; phosphorus, orange). (D,E) Template-directed primer-extension reactions catalyzed by (D) WT 9°N and (E) 9°N-NRQS. Chemical structure of (D) natural phosphodiester-linked DNA and (E) N3′ → P5′ phosphoramidate-linked DNA (NP-DNA).