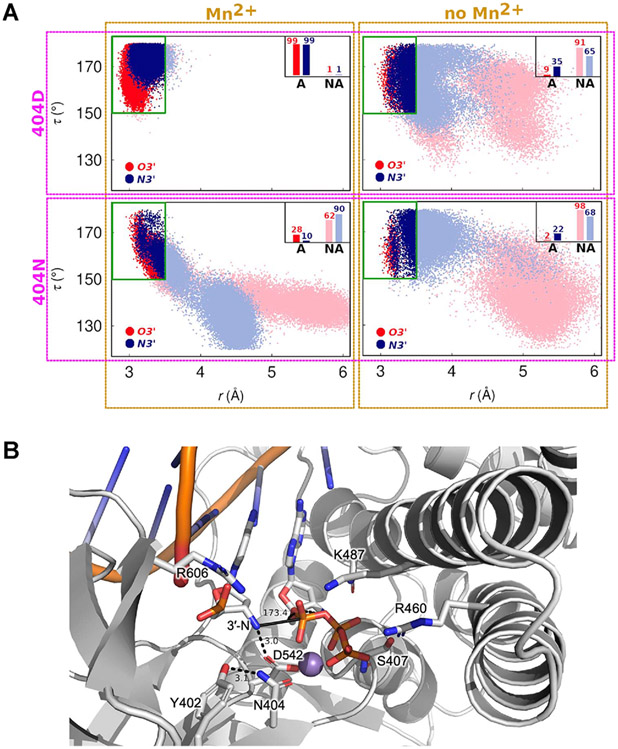
Figure 2.
Analysis of 3′-nucleophile positioning using molecular dynamics (MD) simulations. (A) 2D scatter plots of the in-line attack angle (τ, the angle between the 3′-nucleophile, scissile phosphorus atom, and O5′) and the distance between the 3′-nucleophile and scissile phosphorus atom (r), obtained from MD simulations of 9°N in various active site configurations. In total, eight different active site scenarios are explored: the four panels correspond to simulations in which residue 404 is Asp and the A-site metal ion is present (top left) or absent (top right), residue 404 is Asn and the A-site metal ion is present (bottom left) or absent (bottom right). In each panel, the red and blue colors correspond to the primer being 3′-OH (red) or 3′-NH2 (blue). The data points are classified as active (A) or non-active (NA), where active configurations are defined as those in which the active site possesses in-line fitness (τ > 150°) and in which the 3′-nucleophile is favorably positioned for catalysis (distance between 3′-nucleophile and P ≤ 3.5 Å). The inset bars represent percentage of favorable configurations of the 3′-nucleophile for catalysis via in-line attack. (B) MD snapshot of the 9°N-N active site showing a representative configuration with high in-line fitness (τ ~ 173°, solid black line). The distance between the 3′-amino nucleophile and D542 is 3.0 Å, suggesting that D542 may serve as a base. Observed hydrogen bond and charge interactions in the active site are highlighted (residues in sticks).