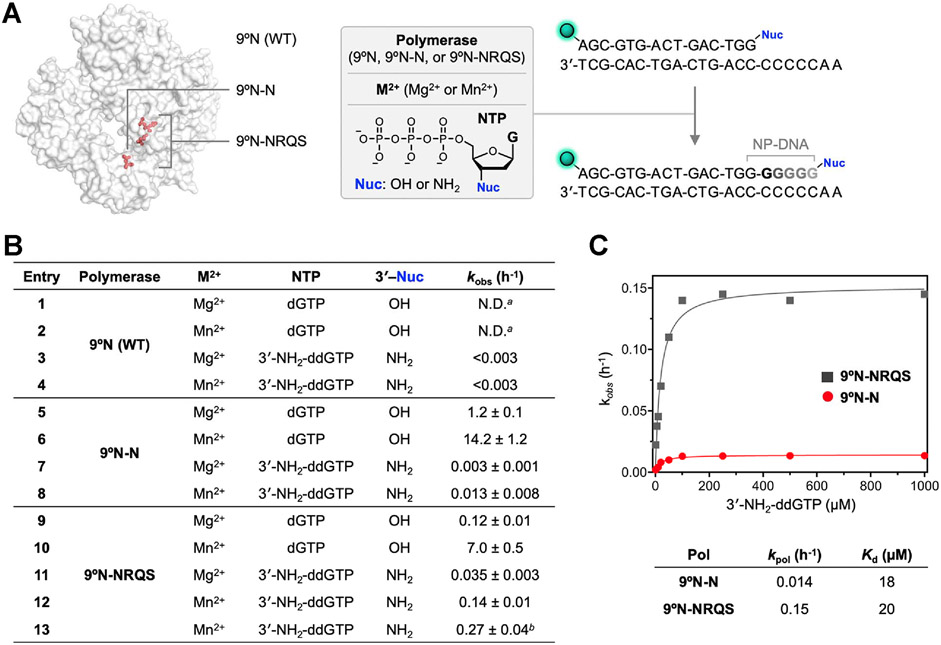
Figure 4.
Enzymatic DNA and NP-DNA primer-extensions. (A) Primer-extension reaction conditions: 1 μM 3′-amino-G primer, 1 μM DNA template, 100 μM 3′-NH2-ddNTP, 1 mM M2+ (Mg2+ or Mn2+), 1 μM polymerase (9°N, 9°N-N, or 9°N-NRQS) in 1× ThermoPol buffer at pH 8.8 at 55 °C. 5′-end of the primer strand is labeled with 6-carboxyfluorescein (FAM), represented as the green sphere. (B) Summary of primer-extension reaction rate, kobs (h−1). For entries 3 and 4, an upper bound of the kobs is provided. a: N.D.: not determined. Product formation is complete in less than 1 min. For 9°N-catalyzed dGTP addition in the presence of Mg2+, kpol was measured as 183,600 h−1 (see ref 35); b: reaction temperature is 65 °C. For entries 5–13, R2 values for the fits are 0.942, 0.991, 0.967, 0.991, 0.913, 0.997, 0.992, 0.994, and 0.977. (C) Substrate concentration-dependent kobs vs 3′-NH2-ddGTP concentration for 9°N-N (red) and 9°N-NRQS (dark gray) at 55 °C. Comparison of maximum rates of 3′-amino-G incorporation, kpol, and dissociation constants, Kd, for both 9°N-N and 9°N-NRQS.