TABLE 2.
Probing promoters cnrYXHa
| Plasmid | Cloned fragment | Maximum fold induction with nickelbd | Transcription | Relative signal strength (%)cd |
|---|---|---|---|---|
| pMOL1551 (1–2418) | 1,027 (11) | Inducible | 100 (0.7) | |
| pMOL1550 (1–2298) | 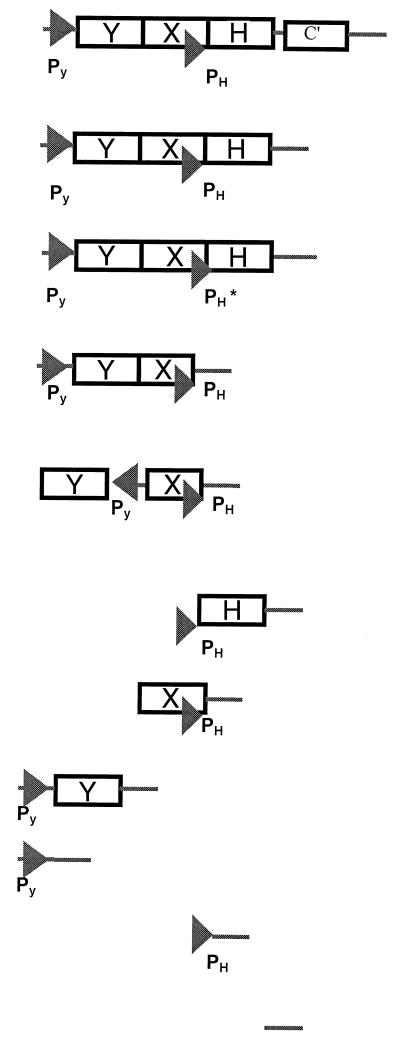 |
1,036 (25) | Inducible | 100 |
| pMOL1561 (1–2298) | <2 | Constitutive | 13 (2.0) | |
| pMOL1586 (870–1720) | 210 (12) | Inducible | 23 (3.5) | |
| pMOL1593 (843–1256), (1256–1720) | <2 | Constitutive | 15 (3.8) | |
| pMOL1588 (1579–2298) | <2 | Constitutive | 58 (9.2) | |
| pMOL1591 (1253–1720) | <2 | Constitutive | 17 (2.8) | |
| pMOL1592 (870–1280) | <2 | Constitutive | 11 (0.7) | |
| pMOL1583 (870–1000) | <2 | Constitutive | 13 (2.8) | |
| pMOL1587 (1579–1720) | <2 | Constitutive | 17 (3.5) | |
| pMOL1596 (2280–2418) | <2 | Constitutive | 0.4 (0.07) | |
| pMOL877 | 0 | Inactive | 0.0 |
Transcriptional fusions between different regions of the cnr regulatory locus and the luxCDABE genes were tested for their metal-responsive activity in vivo in CH34.
The metal response to 0.3 mM nickel was quantitated as the fold induction compared to noninduced cells and is expressed as the signal-to-noise ratio. A value of <2 is considered constitutive transcription.
Maximum light production (in ALU) divided by the OD660nm of the culture, as compared to the transcription of the complete cnr regulatory locus from pMOL1550.
Standard deviations are indicated in parentheses and are based on the results from three independent experiments.
