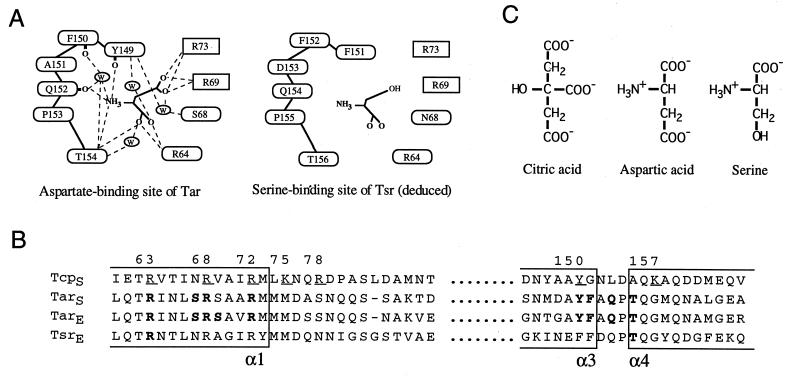
FIG. 1.
Ligand-binding sites of the bacterial chemoreceptors. (A) Schematic illustration of the ligand-binding residues of Tar and Tsr. The hydrogen bonding pattern of the aspartate binding site of Tar was drawn according to Yeh et al. (40). It should be noted that the backbone carbonyl atoms of Gln-152, Phe-150, and Tyr-149 are involved in hydrogen bonding. The serine-binding site of Tsr was deduced from the sequence similarity with Tar. Residues shown in ovals are from one subunit and those in boxes are from the other. W, a bound water molecule. (B) Alignment of the amino acid sequences of the bacterial chemoreceptors of E. coli (indicated with the subscript E) and Salmonella serovar Typhimurium (indicated with the subscript S). Bold letters indicate the residues involved in ligand recognition in Tar and Tsr. Underlined residues of Tcp indicate those replaced by Cys in this study. Numbering of residues is for Tcp. (C) Structures of the attractants specific for Tar, Tsr, or Tcp. The attractants are shown in their fully charged forms.