FIGURE 3.
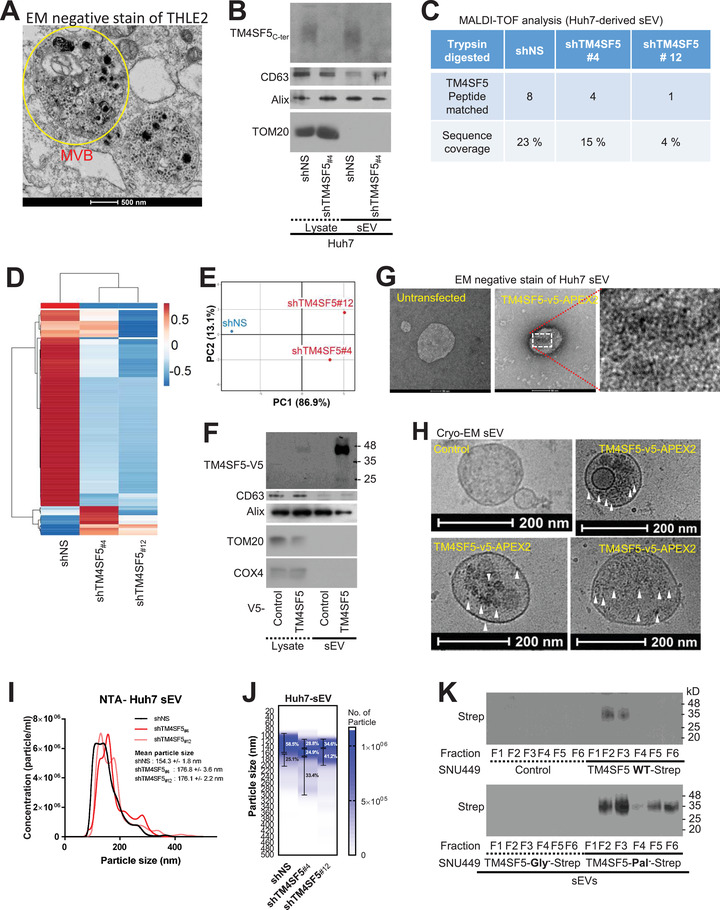
TM4SF5 is recruited into hepatic sEVs. (A) TEM image of Huh7 cells transfected with TM4SF5‐APEX2. Multi‐vesicular body, MVB. (B–E) Western blot analysis for sEV markers and control (B), MALDI‐TOF peptide peak sequence alignment (C), heatmap of common peptide peak intensity profiles (D), and PCA (E) of sEVs from control and TM4SF5‐suppressed Huh7 cells. (F–H) Western blots for v5, sEV markers and controls (F), negative‐stain TEM images (G) and cryo‐EM (H) of sEVs from Huh7 cells transfected with TM4SF5‐APEX2. (I, J) Histogram (I) and heatmap with particle diameters of certain ranges in percentages (J) of sEVs from Huh7 cells stably transfected with shNS or shTM4SF5. (K) SNU449 cells expressing control vector (Cont), Strep‐tagged TM4SF5 WT, N‐glycosylation‐deficient (N138A/N155Q, Gly−) mutant or palmitoylation‐deficient (C2/6/9/74/75/79/80/84/189A, Pal−) mutant were used for sEVs and blotted with Strep‐MAB‐classic‐HRP antibody. The data shown represent three isolated experiments. See also Figure S3
