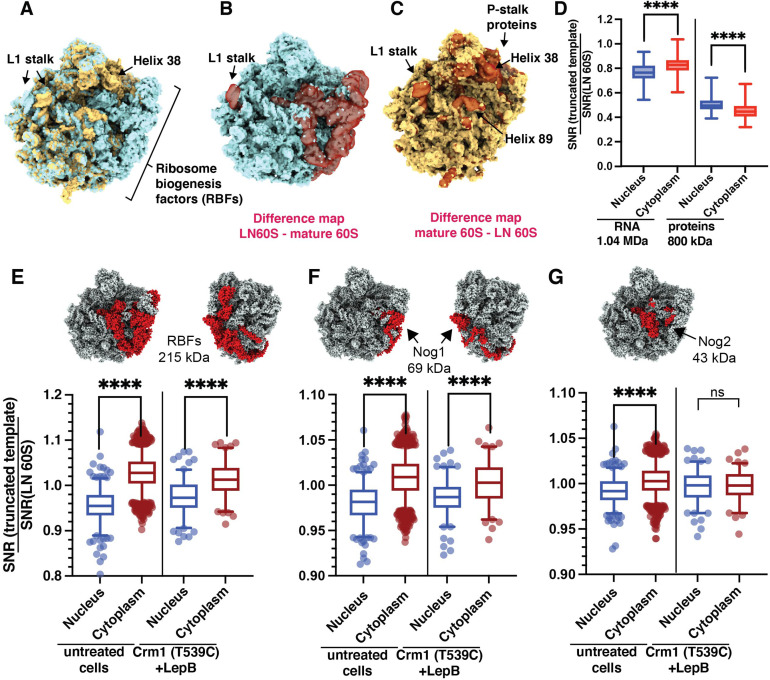
Figure 5. Classification of cytoplasmic mature 60S and nuclear pre-60S by 2DTM corresponds with biologically relevant differences in the templates.
(A) The LN 60S (blue) and mature 60S (yellow) 2DTM templates aligned in UCSF Chimera. (B) LN 60S with difference map calculated using UCSF Chimera showing the density in the LN 60S template that is not present in the mature 60S template (red, transparent). (C) As in (B), showing the mature 60S with density that is not in common with the LN 60S template (red, transparent). (D) Boxplots showing the change in 2DTM SNR when only RNA (left) or protein (right) components of the LN 60S template are included, relative to the full-length template for each significant target. The targets are grouped by their subcellular localization. (E) Upper: LN 60S template with all ribosome biogenesis factors (RBFs) indicated in red. Lower: Boxplot showing the change in the 2DTM SNR of the nuclear (blue) and cytoplasmic (red) targets when all RBFs are removed, relative to the full-length LN 60S template in untreated cells, and when Crm1-mediated nuclear export is inhibited by treating Crm1 (T539C) cells with Leptomycin B (LepB). Box width indicates the interquartile range, the central line indicates the median and the whiskers indicate the range of 95% of the targets. (F) As in (E), for RBF Nog1. (G) As in (E), for RBF Nog2. ****: p<0.0001, ns: not significant (p>0.05).