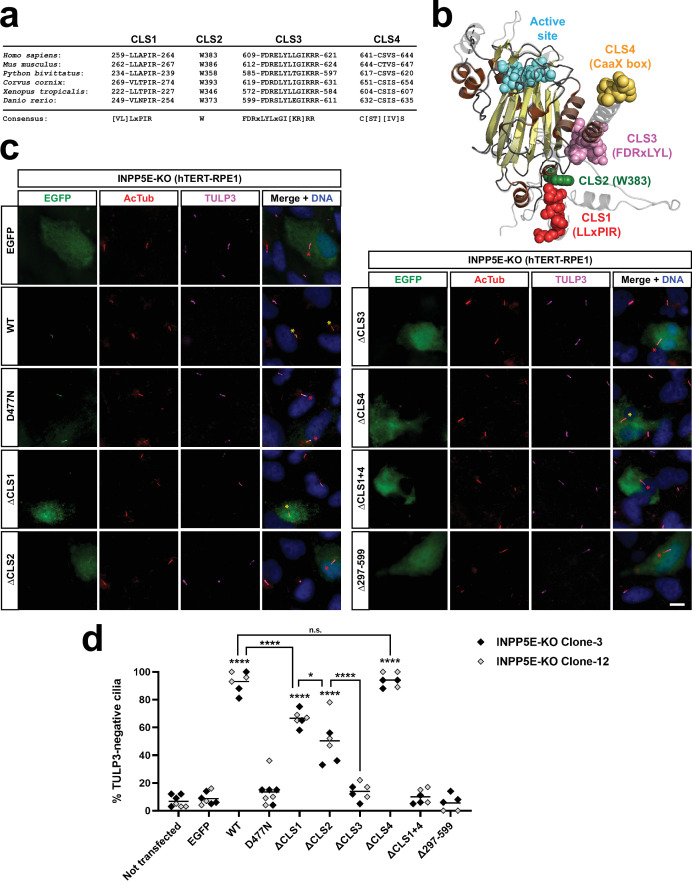
Figure 4. CLS1-4 are conserved ciliary localization signals affecting INPP5E function.
(a) CLS1-4 are highly evolutionarily conserved in vertebrates, including human (NP_063945.2), mouse (AAH80295.1), python (XP_007441606.1), crow (XP_039417670.1), toad (XP_002935265.1), and zebrafish (NP_001096089.2). Consensus sequences are shown below. (b) AlphaFold model of INPP5E 3D structure (AF-Q9NRR6-F1) depicting predicted locations of CLS1 (red), CLS2 (green), CLS3 (pink) and CLS4 (yellow). Active site in cyan. Beta-strands and alpha-helices in yellow and brown, respectively. Proline-rich N-terminal region (aa 1–200), predicted to be highly flexible, is not shown. CLS1 is probably also part of a flexible region, and its position in the AlphaFold model has a low confidence score (pLDDT). See Uniprot entry Q9NRR6 for more details. (c) Rescue assay assessing the ability of INPP5E or its mutants to lower the abnormally high TULP3 levels characteristic of INPP5E-KO cilia. The indicated constructs were transfected into INPP5E-KO RPE1 cells, generated via CRISPR-Cas9 (Figure 4—figure supplement 2). Cells were fixed and stained for EGFP, acetylated tubulin (AcTub), TULP3, and DNA (DAPI), as indicated. Scale bar, 10 µm. Note how untransfected INPP5E-KO cells have high ciliary TULP3 levels, as previously described. Transfected cell cilia are labeled with asterisks in the merge panels: yellow asterisks for rescued TULP3-negative cilia, and red asterisks for non-rescued TULP3-positive cilia. (d) Quantitation of the rescue experiment shown in (c). For each construct, the percentage of TULP3-negative transfected-cell cilia was counted. Data come from five independent experiments. Each point in the graph indicates an independent transfection. Between 12 and 39 transfected-cell cilia were counted per transfection (with exception of the highest data point in ΔCLS2, where only 9 cilia could be counted). Experiments were performed in parallel with two different INPP5E-KO clones (clones 3 and 12). Graph shows individual data points, color-coded by clone as indicated, and the overall median is indicated with a line. Two-way ANOVA revealed significant differences between constructs (p<0.0001) but no significant differences between the clones. All data were then analyzed by one-way ANOVA followed by Tukey tests. Significance is shown relative to EGFP unless otherwise indicated. p<0.0001 (****); p<0.05 (*); not significant (n.s.).