Figure 6. SLC4A7 is required for tumor growth and de novo nucleotide synthesis in tumors.
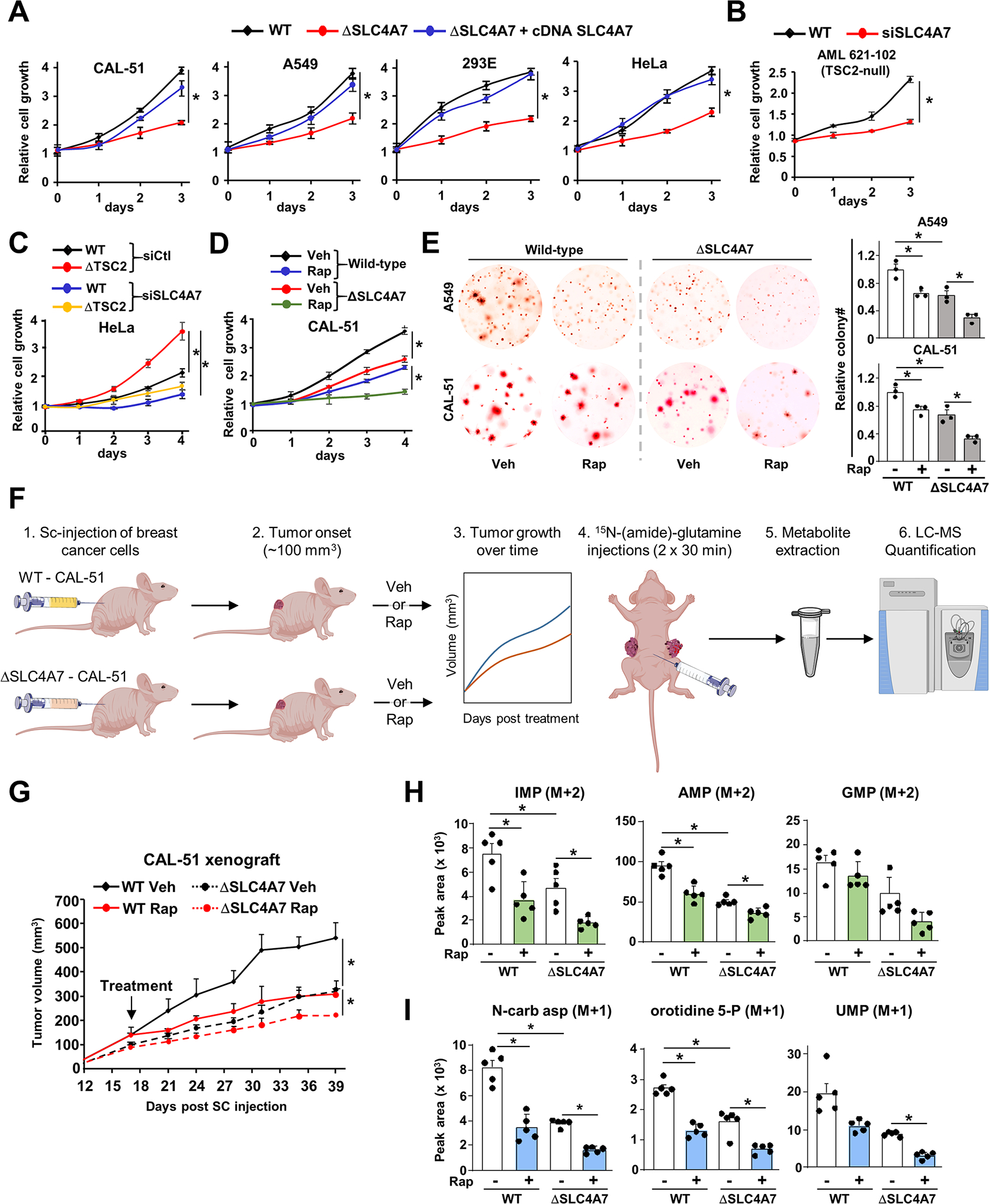
(A) Indicated wild-type or ΔSLC4A7 cell lines were transfected with either empty vector or SLC4A7 cDNA for 48 h and grown in 1 % serum (CAL-51 and A549 cells) or 10 % dialyzed serum (HEK293E and HeLa cells) for 72 h, and cell number was measured via crystal violet staining every 24 h.
(B, C) AML 621–102 (TSC2−/− cells) (B), wild-type and ΔTSC2 HeLa cells (C) grown in 1 % serum for 72 h (B) or 96 h (C), and cell number was measured via crystal violet staining every 24 h.
(D) Wild-type and ΔSLC4A7 CAL-51 cells treated with vehicle or rapamycin (20 nM) for 96 h, and cell number was measured via crystal violet staining every 24 h.
(E) Soft agar colony formation assay with indicated cells treated with vehicle (DMSO) or rapamycin (10 nM) for three weeks. Cell images were acquired at 3× magnification. Colony quantification from three independent biological replicates is shown.
(F) Workflow for the xenograft and in vivo 15N-(amide)-glutamine stable isotope tracing experiments.
(G) Wild-type and ΔSLC4A7 CAL-51 cells were subcutaneously injected into athymic nude mice (n = 5 per group). Mice were treated with vehicle or rapamycin (1 mg/kg), and tumor volume was monitored over time.
(H, I) Mice bearing wild-type or ΔSLC4A7 CAL-51-derived tumors (n = 5) injected with 15N-glutamine (0.8 g/kg) through the intraperitoneal route for 60 min prior to tumor collection and metabolite extraction and analysis via LC-MS/MS.
(A)-(E) The data are plotted as the means ± SDs of biological triplicates and are representative of at least two independent experiments. *p < 0.05, by two-tailed Student’s t-test for pairwise comparisons (B) or one-way ANOVA with Tukey’s post hoc test for multiple pairwise comparisons (A), (C)-(E), (G)-(I).
