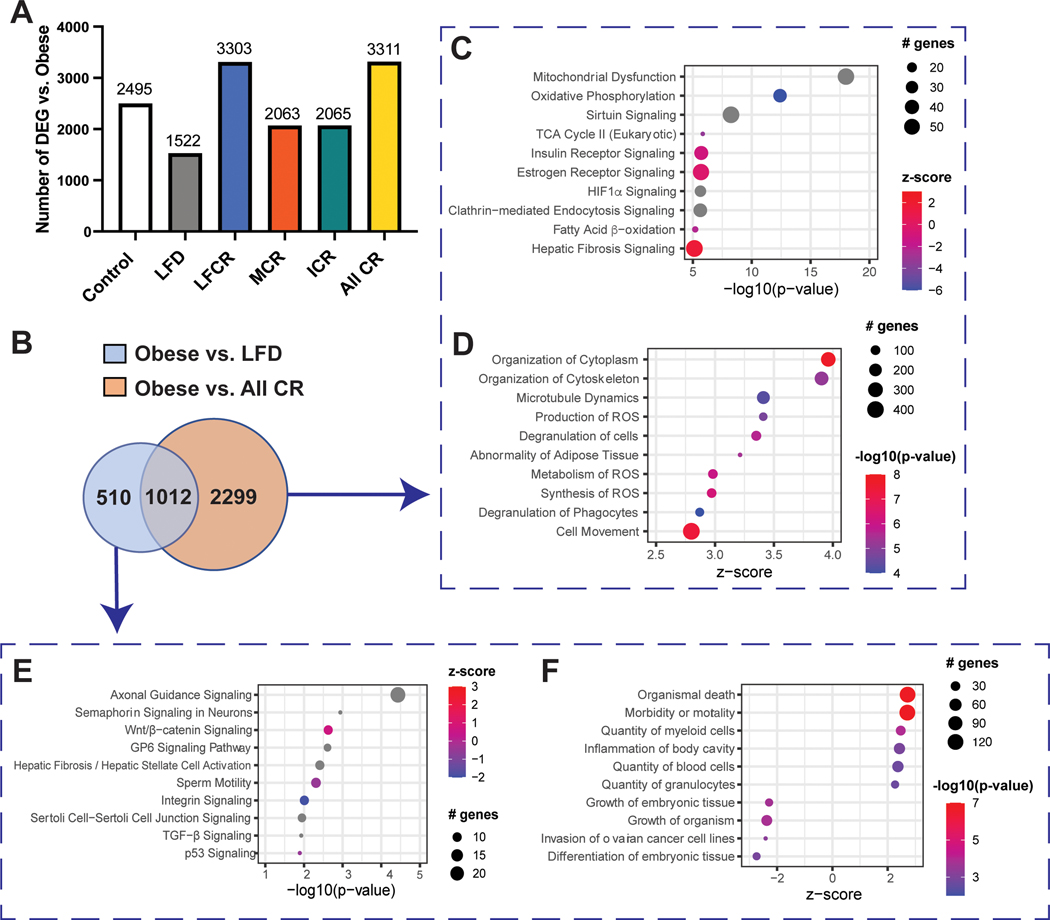
Figure 4.
Calorie restriction modulates gene expression in normal mammary tissue. (A) Differentially expressed genes (DEG, FDR adjusted P<0.05) were identified by RNAseq for obese mice versus every other group (control, nonrestricted low-fat diet (LFD), low-fat calorie-restricted (LFCR), Mediterranean-style calorie-restricted (MCR), and intermittent calorie-restricted (ICR) mice, n=3–5 mice/group) and all calorie-restricted (CR) mice combined. (B) Overlapping and non-overlapping DEG for the obese versus nonrestricted LFD and obese versus all-CR comparisons were determined. (C) The top ten Ingenuity Pathway Analysis (IPA) canonical pathways (ranked by -log10(p-value)) and (D) the top ten significant IPA functions (ranked by z-score) associated with obese versus all-CR DEG, excluding obese versus nonrestricted LFD DEG, are shown. (E) The top ten IPA canonical pathways (ranked by -log10(p-value)) and (F) the top ten significant IPA functions (ranked by z-score) associated with the obese versus nonrestricted LFD DEG, excluding obese versus all-CR DEG, are shown.