Abstract
Background
Procedure-specific complications can have devastating consequences. Machine learning-based tools have the potential to outperform traditional statistical modeling in predicting their risk and guiding decision-making. We sought to develop and compare deep neural network (NN) models, a type of machine learning, to logistic regression (LR) for predicting anastomotic leak after colectomy, bile leak after hepatectomy, and pancreatic fistula after pancreaticoduodenectomy (PD).
Methods
The colectomy, hepatectomy, and PD National Surgical Quality Improvement Program (NSQIP) databases were analyzed. Each dataset was split into training, validation, and testing sets in a 60/20/20 ratio, with 5-fold cross-validation. Models were created using NN and LR for each outcome. Models were evaluated primarily with area under the receiver operating characteristic curve (AUROC).
Results
197,488 patients were included for colectomy, 25,403 for hepatectomy, and 23,333 for PD. For anastomotic leak, AUROC for NN was 0.676 (95% 0.666–0.687), compared with 0.633 (95% CI 0.620–0.647) for LR. For bile leak, AUROC for NN was 0.750 (95% CI 0.739–0.761), compared with 0.722 (95% CI 0.698–0.746) for LR. For pancreatic fistula, AUROC for NN was 0.746 (95% CI 0.733–0.760), compared with 0.713 (95% CI 0.703–0.723) for LR. Variables related to intra-operative information, such as surgical approach, biliary reconstruction, and pancreatic gland texture were highly important for model predictions.
Discussion
Machine learning showed a marginal advantage over traditional statistical techniques in predicting procedure-specific outcomes. However, models that included intra-operative information performed better than those that did not, suggesting that NSQIP procedure-targeted datasets may be strengthened by including relevant intra-operative information.
Keywords: anastomotic leak, pancreatic fistula, hepatectomy, artificial intelligence, machine learning
Introduction
Procedure specific complications can have devastating consequences. For example, anastomotic leak after colectomy is associated with increased morbidity, length of stay, re-admissions, and mortality, as well as local recurrence and cancer-specific mortality for oncologic surgeries.1–3 Predictive models can be helpful to estimate a patient’s specific risk for post-operative complications, guide peri-operative decision-making such as ostomy placement or early drain removal, and perform risk adjustment for comparing post-operative outcomes.
Prior predictive models, such as the American College of Surgeons (ACS) Surgical Risk Calculator, provide accurate estimates of overall mortality and morbidity.4 However, this model, and others which are based on the National Surgical Quality Improvement Program (NSQIP) dataset, fall short in their ability to predict procedure-specific outcomes.5–7
Machine learning, a branch of artificial intelligence (AI), uses computer algorithms that identify patterns within data without explicit instructions and has the potential to identify subtle, non-linear patterns. Machine learning has been successfully applied to the prediction of post-operative outcomes, but previous projects have focused on broader, rather than procedure specific, outcomes, such as overall morbidity and mortality.8,9 Our hypothesis is that machine learning could be helpful in the prediction of procedure-specific outcomes. This study seeks to develop machine learning models for predicting three procedure-specific outcomes: anastomotic leak following colectomy, bile leak following hepatectomy, and pancreatic fistula following pancreaticoduodenectomy (PD). We also sought to compare the machine learning models with logistic regression.
Materials and Methods
Data Source
We used the colectomy, hepatectomy, and pancreatectomy procedure-targeted datasets from the ACS National Surgical Quality Improvement Program (NSQIP) database. All available years for colectomy (2012–2019), hepatectomy (2014–2019), and pancreatectomy (2014–2019) were included. Patients missing primary outcome data were excluded. Patients undergoing colectomy who underwent concurrent ostomy placement were also excluded. From the pancreatectomy dataset, patients undergoing procedures other than PD were excluded. This study was determined to be exempt from institutional review board approval.
Outcomes
For each procedure type, we sought to predict a procedure-specific outcome: anastomotic leak for colectomy, bile leak for hepatectomy, and pancreatic fistula for PD. Anastomotic leak included leaks requiring treatment with antibiotics, percutaneous drainage, or reoperation. Bile leak included leaks requiring percutaneous drainage or reoperation. Pancreatic fistula included grade B or C fistulas for 2018–2019 (fistula grading was implemented in NSQIP in 2018). For 2014–2017, clinically-relevant pancreatic fistulas were defined according to methods described by Kantor et al.6,10
Predictive Models
Each dataset was split into training, validation, and testing sets in 60%, 20%, and 20% ratios, respectively, using randomly selected data from all years. The training set was used for model development, the validation set was used for model adjustment and to monitor overfitting, and the test set was reserved for evaluation of model performance after completion of development. Cross-validation was used to create 5 different train/test splits to verify model consistency. We selected a deep neural network (NN) as our machine learning approach, as it has been previously demonstrated to have improved performance compared with tree-based methods (such as random forest) in prediction of post-operative outcomes from the NSQIP database.8,9,11 This deep learning approach uses layers of functions, each containing model weights, to transform input data into output data representing predictions.12 Dropout (random removal of functions within layers) and early stopping (stopping training when validation set accuracy decreases) were used to reduce overfitting.13 Logistic regression (LR) models were also created for comparison. LR was implemented with no regularization and no variable elimination techniques to approximate a standard implementation. Models were implemented in Python (version 3.9) with use of the Pandas,14,15 SciKitLearn,16 and Keras17 libraries.
Input data included all available peri-operative variables within the core NSQIP database and procedure-targeted variables that would be known prior to the occurrence of the outcome of interest (Tables 1 and 2 and Supplementary Table 1). Missing variables from the datasets were addressed by imputation techniques, which is standard data pre-processing. Missing categorical values were imputed as “unknown” and missing continuous values as the median.9,13,18 Further details are available in the Supplementary Appendix and code is available at https://github.com/gomezlab/nsqip_procedurespecific.
Table 1.
Key Input Variables by Procedure
| Colectomy | Pancreatectomy | Hepatectomy | ||
|---|---|---|---|---|
| Age, mean (SD) | 62.0 (14.9) | 63.4 (12.8) | 59.2 (13.7) | |
| Sex, n (%) | Female | 96357 (53.0) | 19583 (49.8) | 12681 (50.0) |
| Male | 85485 (47.0) | 19711 (50.2) | 12656 (50.0) | |
| Non-binary | 3 (0.0) | 0 (0.0) | 0 (0.0) | |
| Race, n (%) | White | 133433 (73.4) | 29199 (74.3) | 16084 (63.5) |
| Black or African American | 16916 (9.3) | 3327 (8.5) | 2059 (8.1) | |
| Asian | 5571 (3.1) | 1629 (4.1) | 1717 (6.8) | |
| American Indian or Alaska Native | 776 (0.4) | 116 (0.3) | 95 (0.4) | |
| Native Hawaiian or Pacific Islander | 412 (0.2) | 70 (0.2) | 63 (0.2) | |
| Unknown | 24737 (13.6) | 4953 (12.6) | 5319 (21.0) | |
| Hispanic ethnicity n (%) | Yes | 9055 (5.6) | 1977 (5.6) | 1378 (6.7) |
| BMI, mean (SD) | 28.7 (6.7) | 27.9 (6.1) | 28.5 (6.3) | |
| ASA Classification | 1 | 4204 (2.3) | 260 (0.7) | 350 (1.4) |
| 2 | 77345 (42.5) | 9354 (23.8) | 6310 (24.9) | |
| 3 | 87662 (48.2) | 27070 (68.9) | 16800 (66.3) | |
| 4 | 11835 (6.5) | 2552 (6.5) | 1824 (7.2) | |
| 5 | 613 (0.3) | 24 (0.1) | 11 (0.0) | |
| Unknown | 186 (0.1) | 34 (0.1) | 42 (0.2) | |
| Functional Status | Independent | 176926 (97.7) | 38924 (99.2) | 25115 (99.3) |
| Partially Dependent | 3553 (2.0) | 293 (0.7) | 158 (0.6) | |
| Totally Dependent | 679 (0.4) | 25 (0.1) | 15 (0.1) | |
| Dyspnea | At rest | 741 (0.4) | 60 (0.2) | 57 (0.2) |
| With moderate exertion | 11434 (6.3) | 2058 (5.2) | 1337 (5.3) | |
| No | 169670 (93.3) | 37176 (94.6) | 23943 (94.5) | |
| Diabetes | Requiring insulin | 9118 (5.0) | 4839 (12.3) | 1555 (6.1) |
| Not requiring insulin | 18943 (10.4) | 5293 (13.5) | 2938 (11.6) | |
| No diabetes | 153784 (84.6) | 29162 (74.2) | 20844 (82.3) | |
| Hypertension | 87817 (48.3) | 20445 (52.0) | 11589 (45.7) | |
| Heart failure | 1949 (1.1) | 157 (0.4) | 93 (0.4) | |
| Ascites | 996 (0.5) | 114 (0.3) | 131 (0.5) | |
| COPD | 9016 (5.0) | 1586 (4.0) | 900 (3.6) | |
| Renal failure | 697 (0.4) | 30 (0.1) | 22 (0.1) | |
| Dialysis | 1598 (0.9) | 156 (0.4) | 81 (0.3) | |
| Chronic steroid use | 13313 (7.3) | 1220 (3.1) | 817 (3.2) | |
| Smoking | 28987 (15.9) | 6703 (17.1) | 3851 (15.2) | |
| Bleeding disorder | 6593 (3.6) | 1206 (3.1) | 842 (3.3) | |
| Weight loss (>10%) | 7279 (4.0) | 4659 (11.9) | 975 (3.8) | |
| Pre-operative transfusion | 4213 (2.3) | 319 (0.8) | 147 (0.6) | |
| Wound classification | Clean | 1823 (1.0) | 2621 (6.7) | 3647 (14.4) |
| Clean/contaminated | 139733 (76.8) | 31308 (79.7) | 20032 (79.1) | |
| Contaminated | 22625 (12.4) | 4317 (11.0) | 1129 (4.5) | |
| Dirty/Infected | 17664 (9.7) | 1048 (2.7) | 529 (2.1) | |
| Transfer status | Not transferred | 172906 (95.1) | 38211 (97.3) | 24881 (98.2) |
| From acute care hospital | 3632 (2.0) | 781 (2.0) | 277 (1.1) | |
| From nursing home | 1502 (0.8) | 75 (0.2) | 38 (0.2) | |
| From outside ED | 3148 (1.7) | 169 (0.4) | 110 (0.4) | |
| From other | 544 (0.3) | 51 (0.1) | 26 (0.1) | |
| Sodium, mean (SD) | 139.1 (3.1) | 139.0 (3.1) | 139.3 (2.8) | |
| Blood urea nitrogen, mean (SD) | 15.5 (9.5) | 15.6 (7.4) | 15.1 (6.9) | |
| Creatinine, mean (SD) | 1.0 (0.7) | 0.9 (0.5) | 0.9 (0.5) | |
| Albumin, mean (SD) | 3.8 (0.6) | 3.9 (0.6) | 4.0 (0.5) | |
| White blood cell count, mean (SD) | 7.9 (3.6) | 7.3 (2.8) | 6.9 (3.1) | |
| Hematocrit, mean (SD) | 38.3 (5.9) | 38.3 (5.2) | 39.4 (5.0) | |
| Platelet count, mean (SD) | 268.0 (95.3) | 250.0 (91.6) | 236.2 (90.8) | |
| Operative time, mean (SD) | 173.0 (88.2) | 371.9 (128.5) | 239.9 (121.7) |
Data are n (%) unless otherwise specified. BMI = Body Mass Index. ASA = American Society of Anesthesiologists. COPD = Chronic obstructive pulmonary disease. PATOS = present at time of surgery
Table 2.
Procedure-Targeted Variables for Colectomy, Hepatectomy, and Pancreatectomy
| Colectomy | ||
| CPT, n (%) | Colectomy | 28472 (15.7) |
| Colectomy with coloproctostomy | 14051 (7.7) | |
| Colectomy with abdominal and transanal approach | 312 (0.2) | |
| Colectomy with ileocolostomy | 23458 (12.9) | |
| Laparoscopic colectomy | 48250 (26.5) | |
| Laparoscopic colectomy with ileocolostomy | 33206 (18.3) | |
| Laparoscopic colectomy with coloproctostomy | 34096 (18.8) | |
| Indication, n (%) | Acute diverticulitis | 11348 (5.8) |
| Bleeding | 1244 (0.6) | |
| Chronic diverticular disease | 30920 (15.7) | |
| Colon cancer | 75478 (38.4) | |
| Colon cancer w/ obstruction | 8433 (4.3) | |
| Crohn’s Disease | 11641 (5.9) | |
| Enterocolitis (e.g. C. Difficile) | 395 (0.2) | |
| Non-malignant polyp | 18981 (9.7) | |
| Other | 31764 (16.1) | |
| Ulcerative colitis | 846 (0.4) | |
| Volvulus | 5609 (2.9) | |
| Emergent indication, n (%) | Not emergent | 178150 (90.4) |
| Bleeding | 1121 (0.6) | |
| Obstruction | 6904 (3.5) | |
| Other | 2256 (1.1) | |
| Perforation | 6072 (3.1) | |
| Toxic colitis | 948 (0.5) | |
| Pre-operative steroid use, n (%) | 10459 (5.4) | |
| Mechanical bowel prep, n (%) | 109434 (63.9) | |
| Antibiotic bowel prep, n (%) | 81762 (47.1) | |
| Pre-operative chemotherapy, n (%) | 7485 (3.8) | |
| Approach, n (%) | Open (planned) | 55977 (28.4) |
| Laparoscopic | 61348 (31.2) | |
| Laparoscopic w/ open assist | 46797 (23.8) | |
| Laparoscopic w/ unplanned conversion to open | 13803 (7.0) | |
| Robotic | 11531 (5.9) | |
| Robotic w/ open assist | 6283 (3.2) | |
| Robotic w/ unplanned conversion to open | 969 (0.5) | |
| Other | 127 (0.1) | |
| Hepatectomy | ||
| CPT code, n (%) | Hepatectomy, partial lobectomy | 17073 (67.4) |
| Hepatectomy, trisegmentectomy | 2050 (8.1) | |
| Hepatectomy, total left lobectomy | 2274 (9.0) | |
| Hepatectomy, total right lobectomy | 3940 (15.6) | |
| Indication, n (%) | Colorectal metastasis | 8403 (33.1) |
| Other metastasis | 1503 (6.0) | |
| Hepatocellular carcinoma | 4575 (18.0) | |
| Cholangiocarcinoma | 2233 (8.8) | |
| Hepatic adenoma | 1005 (4.0) | |
| Hemangioma | 802 (3.2) | |
| Hepatic cyst | 722 (2.8) | |
| Gallbladder cancer | 655 (2.6) | |
| Focal nodular hyperplasia | 474 (1.9) | |
| Biliary cyst | 416 (1.6) | |
| Hepatic abscess | 190 (0.7) | |
| Other | 4425 (17.4) | |
| Biliary stent placed, n (%) | Yes, endoscopic | 948 (3.8) |
| Yes, percutaneous | 216 (0.9) | |
| Yes, other/unknown | 102 (0.4) | |
| No | 23943 (95.0) | |
| Unknown | 194 (0.8) | |
| Drain placed, n (%) | 11229 (44.3) | |
| Neo-adjuvant systemic chemotherapy, n (%) | 6566 (25.8) | |
| Portal vein embolization, n (%) | 877 (3.5) | |
| Pre-operative intra-arterial infusion, n (%) | 222 (0.9) | |
| Pre-operative ablation, n (%) | 169 (0.7) | |
| Viral hepatitis, n (%) | Hepatitis B | 1124 (4.9) |
| Hepatitis B and C | 133 (0.6) | |
| Hepatitis C | 1670 (7.3) | |
| None | 19677 (86.4) | |
| Other | 158 (0.7) | |
| Approach, n (%) | MIS | 5777 (22.8) |
| MIS w/ conversion | 999 (3.9) | |
| Open (planned) | 18616 (73.3) | |
| Liver texture, n (%) | Cirrhotic | 2461 (9.7) |
| Congested | 468 (1.8) | |
| Fatty | 3229 (12.7) | |
| Fibrosis | 256 (1.0) | |
| Normal | 7030 (27.7) | |
| Unknown | 11959 (47.1) | |
| Number of concurrent partial resections, n (%) | 0 | 12688 (50.7) |
| 1 | 6822 (27.3) | |
| 2 | 3011 (12.0) | |
| 3 or more | 2439 (9.8) | |
| Pancreatectomy | ||
| CPT, n (%) | Pancreaticoduodenectomy | 14679 (63.2) |
| Pylorus-sparing pancreaticoduodenectomy | 8554 (36.8) | |
| Indication, n (%) | Pancreatic adenocarcinoma | 12931 (55.7) |
| Ampullary/duodenal adenocarcinoma | 3627 (15.6) | |
| Biliary adenocarcinoma | 1761 (7.6) | |
| Neuroendocrine tumor | 1247 (5.5) | |
| Benign neoplasm of pancreas | 945 (4.1) | |
| Cystic lesion | 1101 (4.7) | |
| Chronic pancreatitis | 865 (3.7) | |
| Other | 756 (3.3) | |
| Jaundice, n (%) | 10102 (43.8) | |
| Pre-operative biliary stent, n (%) | Endoscopic stent | 10950 (49.1) |
| No stent at time of surgery | 10229 (45.9) | |
| Percutaneous stent | 696 (3.1) | |
| Stent of other or unknown type | 405 (1.8) | |
| Pre-operative chemotherapy, n (%) | 4857 (21.0) | |
| Pre-operative radiation therapy, n (%) | 1863 (8.1) | |
| Approach, n (%) | Minimally invasive (MIS) | 1863 (8.1) |
| Open (planned) | 21172 (91.1) | |
| Pancreatic duct size, n (%) | 3–6 mm | 9780 (42.1) |
| <3 mm | 5748 (24.7) | |
| >6 mm | 3031 (13.0) | |
| Unknown | 4674 (20.1) | |
| Pancreas gland texture, n (%) | Hard | 7517 (32.4) |
| Intermediate | 2117 (9.1) | |
| Soft | 8143 (35.0) | |
| Unknown | 5456 (23.5) | |
| Type of reconstruction, n (%) | Not performed | 739 (3.3) |
| Pancreaticogastrostomy | 511 (2.3) | |
| Pancreaticojejunal duct-to-mucosal | 19499 (86.0) | |
| Pancreaticojejunal invagination | 1915 (8.4) | |
| Drains placed, n (%) | Yes | 20649 (89.0) |
| Vascular resection, n (%) | Not performed | 18950 (82.4) |
| Artery | 435 (1.9) | |
| Vein | 2860 (12.4) | |
| Vein and artery | 766 (3.3) | |
| Drain amylase (POD1), mean (SD) | 3475.8 (10299.8) | |
| Incision type, n (%) | Subcostal type | 1916 (8.2) |
| Upper midline | 9179 (39.5) | |
| Other | 177 (0.8) | |
| Unknown | 11961 (51.5) | |
| Gastrojejunostomy, n (%) | Antecolic | 3832 (16.5) |
| Retrocolic | 1611 (6.9) | |
| Not performed | 192 (0.8) | |
| Unknown | 17598 (75.7) | |
| Drain location, n (%) | Biliary anastomosis | 157 (0.7) |
| Pancreatic & Biliary Anastomosis | 3946 (17.0) | |
| Pancreatic anastomosis | 964 (4.1) | |
| Pancreatic parenchyma | 119 (0.5) | |
| Type(s) cannot be determined | 536 (2.3) | |
| Unknown | 17511 (75.4) | |
| Drain system type, n (%) | Closed | 10599 (45.6) |
| Closed and Open | 122 (0.5) | |
| Open | 96 (0.4) | |
| Unknown | 12416 (53.4) | |
| Wound protector, n (%) | Yes | 4131 (17.8) |
| No | 11334 (48.8) | |
| Unknown | 7768 (33.4) | |
| Pre-incision antibiotic, n (%) | 1st generation cephalosporin | 5302 (22.8) |
| 2nd or 3rd generation cephalosporin | 4493 (19.3) | |
| Broad spectrum | 6125 (26.4) | |
| Other | 552 (2.4) | |
| Unknown | 6761 (29.1) |
Evaluation
Models were evaluated primarily with area under the receiver operating characteristic curve (AUROC). The receiver operating characteristic curve plots the true positive rate against the false positive rate and the AUROC summarizes the model’s ability to distinguish positive cases from negative cases. AUROC ranges from 0.5 (random guessing) to 1 (perfect classification). AUROC’s were compared between models using the Delong test with significance set at p <0.05.19 In addition, the area under the precision-recall curve (AUPRC) was also calculated for each model, which assesses a model’s ability to identify all positive cases without identifying false positives. A random classifier will have an AUPRC equal to the rate of the positive class (e.g., rate of anastomotic leak) and a perfect classifier will have an AUPRC of 1.0. The relative importance of input variables was estimated for procedure-specific variables using Shapley additive explanations (SHAP) for NN models and odds ratios for LR models.20
Results
Colectomy
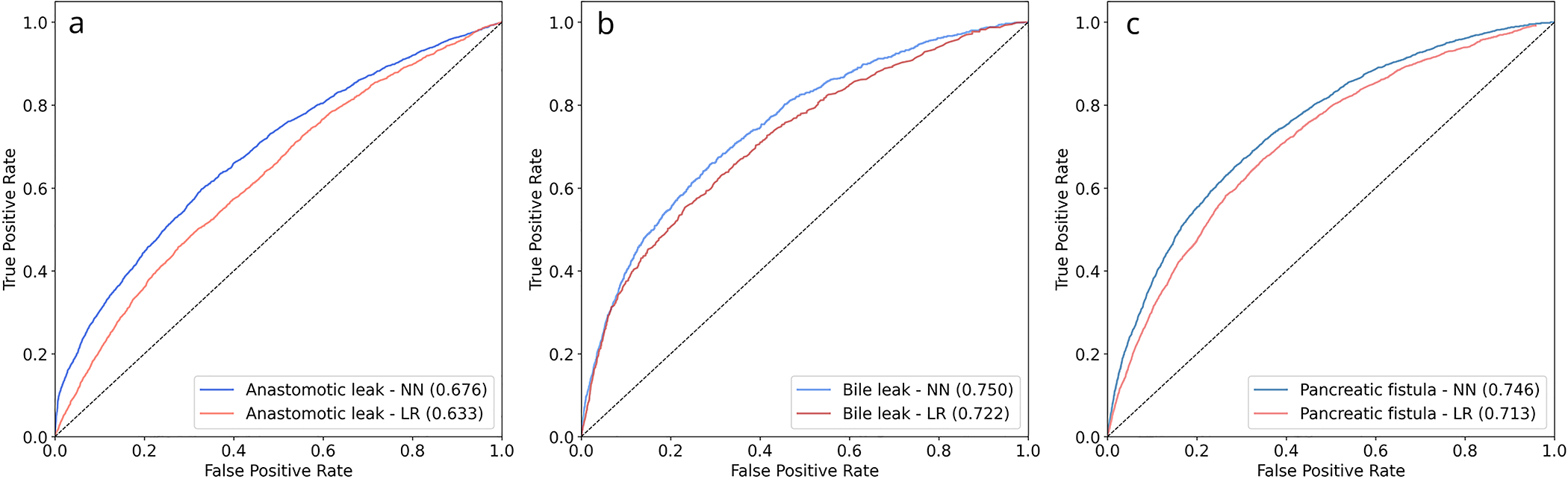
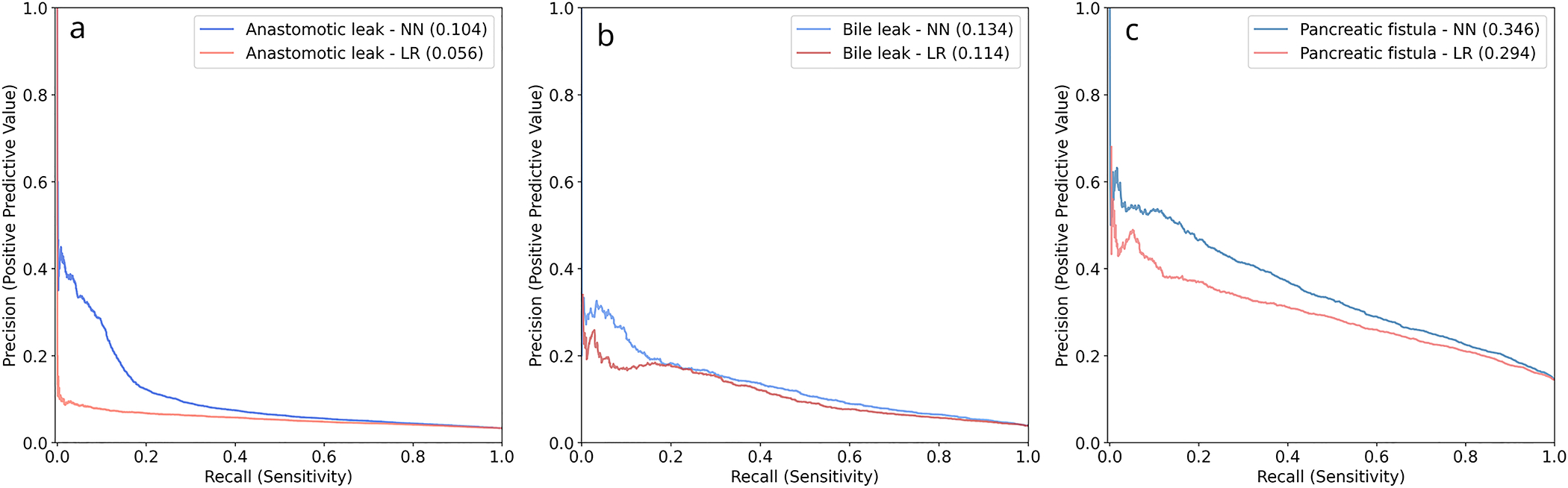
The colectomy dataset included 257,913 patients. After application of exclusion criteria, 197,488 patients remained. 6012 (3.05%) patients experienced an anastomotic leak. After splitting, 118,493 patients were included in the training group, 39,497 patients were included in the validation group, and 39,498 patients were included in the test group. Further input variable characteristics for all groups are described in Table 1. On the test set, NN obtained an AUROC of 0.676 (95% 0.666–0.687) and an AUPRC of 0.104 (95% CI 0.092–0.115). LR obtained an AUROC of 0.633 (95% CI 0.620–0.647) and an AUPRC of 0.056 (95% CI 0.051–0.061). Receiver operating characteristic and precision-recall curves for anastomotic leak are shown in Figures 1a and 2a. Comparison using the Delong test showed a significant difference between the AUROC of NN and LR with p <0.001. Of the variables within the procedure-targeted dataset, approach, mechanic bowel prep, and antibiotic bowel prep contributed most to the NN model output, compared with chemotherapy, pre-operative steroid use, and antibiotic bowel prep for the LR model (Table 4).
Figure 1.
Receiver Operating Characteristic Curves for Procedure-Specific Outcomes, NN = neural network, LR = logistic regression, a = Anastomotic leak, b = Bile leak, c = Pancreatic fistula
Figure 2.
Precision-Recall Curves for Procedure-Specific Outcomes, NN = neural network, LR = logistic regression, a = Anastomotic leak, b = Bile leak, c = Pancreatic fistula
Table 4.
Relative Importance of Input Variables Compared between Neural Network and Logistic Regression Using SHAP Values and Odds Ratios
| Anastomotic leak | |||
| Variable | SHAP value | Variable | Odds ratio* |
|
| |||
| Approach | 0.016 | Chemotherapy | 1.32 |
|
| |||
| Mechanical bowel prep | 0.016 | Steroid use | 1.25 |
|
| |||
| Antibiotic bowel prep | 0.014 | Antibiotic bowel prep | 0.81 |
|
| |||
| Emergent indication | 0.011 | Mechanical bowel prep | 0.86 |
|
| |||
| Steroid use | 0.010 | Approach | 1.14 |
|
| |||
| Chemotherapy | 0.009 | Emergent indication | 0.94 |
|
| |||
| Indication | 0.009 | Indication | 1.01 |
|
| |||
| Bile leak | |||
| Variable | SHAP value | Variable | Odds ratio* |
|
| |||
| Use of drain | 0.034 | Biliary reconstruction | 1.88 |
|
| |||
| Biliary reconstruction | 0.029 | Pringle maneuver | 1.42 |
|
| |||
| Approach | 0.017 | Approach | 1.37 |
|
| |||
| Biliary stent | 0.016 | Neoadjuvant chemo-embolization | 1.37 |
|
| |||
| Pringle maneuver | 0.015 | Use of drain | 1.37 |
|
| |||
| # of concurrent resections | 0.011 | Neoadjuvant chemo-infusion | 0.73 |
|
| |||
| Concurrent ablation | 0.01 | Biliary stent | 1.22 |
|
| |||
| Viral hepatitis | 0.009 | Neoadjuvant ablation | 1.19 |
|
| |||
| Neoadjuvant therapy | 0.009 | Neoadjuvant chemotherapy | 1.17 |
|
| |||
| Neoadjuvant chemo-embolization | 0.008 | Viral hepatitis | 1.13 |
|
| |||
| Pancreatic fistula | |||
| Variable | SHAP value | Variable | Odds ratio* |
|
| |||
| Gland texture | 0.039 | Drains placed | 1.27 |
|
| |||
| Indication | 0.036 | Gland texture | 1.25 |
|
| |||
| Drain amylase (POD1) | 0.027 | Chemotherapy | 0.89 |
|
| |||
| Reconstruction | 0.010 | Reconstruction | 1.09 |
|
| |||
| Duct size | 0.008 | Indication | 0.92 |
|
| |||
| Vascular resection | 0.006 | Radiation therapy | 0.93 |
|
| |||
| Biliary stent | 0.006 | Vascular resection | 0.94 |
|
| |||
| Jaundice | 0.006 | Duct size | 0.94 |
|
| |||
| Radiation therapy | 0.006 | Antibiotic | 0.96 |
|
| |||
| Chemotherapy | 0.005 | Jaundice | 0.97 |
Odds ratio is sorted by distance from 1 (null value)
Hepatectomy
The hepatectomy dataset included 25,595 patients. After application of exclusion criteria, 25,403 patients remained. 966 (3.8%) patients experienced a bile leak. After splitting, 15,242 patients were included in the training group, 5,080 patients were included in the validation group, and 5,081 patients were included in the test group. On the test set, NN obtained an AUROC of 0.750 (95% CI 0.739–0.761) and an AUPRC of 0.134 (95% CI 0.115–0.153). LR obtained an AUROC of 0.722 (95% CI 0.698–0.746) and AUPRC of 0.114 (95% CI 0.090–0.139). Receiver operating characteristic and precision-recall curves for anastomotic leak are shown in Figures 1b and 2b. Comparison using the Delong test showed a significant difference between the AUROC of NN and LR with p = 0.003. Of the variables within the procedure-targeted dataset, placement of drain intra-operatively, biliary reconstruction, surgical approach, biliary stent placement, use of Pringle maneuver, and number of concurrent resections contributed most to the NN model, compared with biliary reconstruction, Pringle maneuver, surgical approach, neoadjuvant chemo-embolization, placement of drain, and neoadjuvant chemo-infusion for the LR model (Table 4).
Pancreaticoduodenectomy
The PD dataset included 23,437 patients. After application of exclusion criteria, 23,233 patients remained. 3,346 (14.4%) patients experienced a pancreatic fistula. After splitting, 13,940 patients were included in the training group, 4,647 patients were included in the validation group, and 4,646 patients were included in the test group. On the test set, NN obtained an AUROC of 0.746 (95% CI 0.733–0.760) and an AUPRC of 0.346 (95% CI 0.327–0.365). LR obtained an AUROC of 0.713 (95% CI 0.703–0.723) and an AUPRC of 0.294 (95% CI 0.281–0.307). Receiver operating characteristic and precision-recall curves for anastomotic leak are shown in Figures 1c and 2c. Comparison using the Delong test showed a significant difference between the AUROCs of NN and LR with p < 0.001. Of the variables within the procedure-targeted dataset, pancreatic gland texture, indication, drain amylase on post-operative day 1, type of reconstruction, and duct size contributed most to the NN model output, compared with placement of drain intra-operatively, gland texture, pre-operative chemotherapy, type of reconstruction, and indication for the LR model (Table 4).
Discussion
This study developed and compared machine learning and logistic regression models which predict procedure-specific complications after colectomy, hepatectomy, and PD. Overall, the NN showed marginal improvement over LR in terms of predictive accuracy. There was a marked difference between models’ predictive ability for various outcomes, with anastomotic leak after colectomy less accurately predicted compared with bile leak after hepatectomy and pancreatic fistula after PD for both the NN and LR approaches. Evaluation of variable importance using SHAP values and odds ratios showed that both models emphasized intra-operative variables as risk factors. Notably, the colectomy procedure-targeted dataset includes much less intra-operative information compared with hepatectomy and PD.
While machine learning applied to the entire NSQIP dataset predicts general outcomes with high accuracy (AUROC 0.88–0.95) and significantly outperforms the ACS risk calculator,4,8 machine learning to predict procedure-specific complications in the current project does not show as clear of an advantage over LR. For anastomotic leak, previous models developed using LR and the NSQIP dataset obtained AUROC’s of 0.65–0.66, similar to our machine learning models, although they significantly outperform the ACS Surgical Risk Calculator (AUROC 0.58).5,21,22 Models developed using LR on single-institution and regional datasets, which also incorporate more intra-operative information, have obtained higher AUROC’s 0.73 – 0.82.7,23 LR models created for bile leak and pancreatic leak from non-NSQIP datasets resulted in AUROC (0.65–0.79), similar to results for our models.24–30 One previous study did apply machine learning methods to predict pancreatic fistula in a smaller, single-institution dataset of 1769 patients with an AUROC 0.74, also similar to our model.31
A particularly interesting finding from this study is that certain outcomes, in particular anastomotic leak after colectomy, are much more difficult to predict from the NSQIP dataset compared with bile leak and pancreatic fistula. This is likely because the NSQIP dataset does not include intra-operative variables for colectomy, in contrast to hepatectomy and pancreatectomy. Tellingly, models for anastomotic leak based on non-NSQIP datasets which include relevant intra-operative information, such as number of staple fires, occurrence of intra-operative adverse events, and need for intra-operative transfusion have improved accuracy (AUROC 0.73 – 0.82) that are more similar our results for hepatectomy and PD.7,23 This aligns with a body of literature showing a strong link between intra-operative performance and post-operative outcomes, indicating that the incorporation of intra-operative information is key to predicting procedure-specific outcomes.31–34 32–34
This comparison does have some limitations. First, use of NSQIP as training data introduces selection bias because only hospitals participating in the NSQIP program are included. In addition, predictions are limited to 30-day outcomes. For some variables, data may be missing because of the clinical scenario and for those variables, assumptions made using imputation techniques may not be valid. Missing data for pancreatectomy variables has also improved over time, making earlier years less useful for model training. Second, this study is not an exhaustive analysis of every procedure-specific complication in NSQIP. Rather, it analyzes the abdominal surgical procedures with the most robust procedure-targeted datasets. Finally, while direct comparison of the absolute values of SHAP and odds ratios is not valid, their use for relative importance can provide insights into model decision-making.
Conclusion
In conclusion, our results show that machine learning has a marginal advantage over traditional statistical techniques in predicting procedure-specific outcomes based on the NSQIP dataset. However, models which include intra-operative variables performed better compared with those that did not, suggesting that NSQIP procedure-targeted datasets may be strengthened by the collection of relevant intra-operative information. The application of machine learning to datasets which include multi-modal data, such as real-time electronic health record information and assessments of intra-operative surgeon performance, represents a target of future research.
Supplementary Material
Table 3.
Area Under the Receiver Operating Characteristic and Precision-Recall Curves for Neural Network and Logistic Regression Models
| AUROC Mean | AUROC 95% CI | AUPRC Mean | AUPRC 95% CI | |
|---|---|---|---|---|
| Anastomotic Leak - NN | 0.68 | 0.67–0.69 | 0.10 | 0.09–0.12 |
| Anastomotic Leak - LR | 0.63 | 0.62–0.65 | 0.06 | 0.05–0.06 |
| Bile Leak - NN | 0.75 | 0.74–0.76 | 0.13 | 0.12–0.15 |
| Bile Leak - LR | 0.72 | 0.70–0.75 | 0.11 | 0.10–0.14 |
| Pancreatic Fistula - NN | 0.75 | 0.73–0.76 | 0.35 | 0.33–0.37 |
| Pancreatic Fistula - LR | 0.71 | 0.70–0.72 | 0.29 | 0.28–0.30 |
Acknowledgments
Grant support: This work was supported by funding from the National Institutes of Health (Program in Translational Medicine T32-CA244125 to UNC/KAC).
Footnotes
Conflicts of interest: None declared for each author
References
- 1.Midura EF, Hanseman D, Davis BR, et al. Risk factors and consequences of anastomotic leak after colectomy: A national analysis. In: Diseases of the Colon and Rectum. Vol 58. Lippincott Williams and Wilkins; 2015:333–338. doi: 10.1097/DCR.0000000000000249 [DOI] [PubMed] [Google Scholar]
- 2.Mirnezami A, Mirnezami R, Chandrakumaran K, Sasapu K, Sagar P, Finan P. Increased local recurrence and reduced survival from colorectal cancer following anastomotic leak: Systematic review and meta-analysis. Ann Surg. 2011;253(5):890–899. doi: 10.1097/SLA.0b013e3182128929 [DOI] [PubMed] [Google Scholar]
- 3.Romagnoni A, Jégou S, Van Steen K, et al. Comparative performances of machine learning methods for classifying Crohn Disease patients using genome-wide genotyping data. Sci Reports 2019 91. 2019;9(1):1–18. doi: 10.1038/s41598-019-46649-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.KY B, Y L, JL P, et al. Development and evaluation of the universal ACS NSQIP surgical risk calculator: a decision aid and informed consent tool for patients and surgeons. J Am Coll Surg. 2013;217(5). doi: 10.1016/J.JAMCOLLSURG.2013.07.385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McKenna NP, Bews KA, Cima RR, Crowson CS, Habermann EB. Development of a Risk Score to Predict Anastomotic Leak After Left-Sided Colectomy: Which Patients Warrant Diversion? J Gastrointest Surg. 2020;24(1):132–143. doi: 10.1007/s11605-019-04293-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kantor O, Talamonti MS, Pitt HA, et al. Using the NSQIP Pancreatic Demonstration Project to Derive a Modified Fistula Risk Score for Preoperative Risk Stratification in Patients Undergoing Pancreaticoduodenectomy. J Am Coll Surg. 2017;224(5):816–825. doi: 10.1016/j.jamcollsurg.2017.01.054 [DOI] [PubMed] [Google Scholar]
- 7.T S, L C, AI K, et al. Validation of an online risk calculator for the prediction of anastomotic leak after colon cancer surgery and preliminary exploration of artificial intelligence-based analytics. Tech Coloproctol. 2017;21(11):869–877. doi: 10.1007/S10151-017-1701-1 [DOI] [PubMed] [Google Scholar]
- 8.Bertsimas D, Dunn J, Velmahos GC, Kaafarani HMA. Surgical Risk Is Not Linear: Derivation and Validation of a Novel, User-friendly, and Machine-learning-based Predictive OpTimal Trees in Emergency Surgery Risk (POTTER) Calculator. Ann Surg. 2018;268(4):574–583. doi: 10.1097/SLA.0000000000002956 [DOI] [PubMed] [Google Scholar]
- 9.Varadarajan KM, Muratoglu OK, Malchau H, et al. Assessing the utility of deep neural networks in predicting postoperative surgical complications: a retrospective study. Artic Lancet Digit Heal. 2021;3:471–485. doi: 10.1016/S2589-7500(21)00084-4 [DOI] [PubMed] [Google Scholar]
- 10.C B, G M, C D, et al. The 2016 update of the International Study Group (ISGPS) definition and grading of postoperative pancreatic fistula: 11 Years After. Surgery. 2017;161(3):584–591. doi: 10.1016/J.SURG.2016.11.014 [DOI] [PubMed] [Google Scholar]
- 11.Merath K, Hyer JM, Mehta R, et al. Use of Machine Learning for Prediction of Patient Risk of Postoperative Complications After Liver, Pancreatic, and Colorectal Surgery. J Gastrointest Surg 2019 248. 2019;24(8):1843–1851. doi: 10.1007/S11605-019-04338-2 [DOI] [PubMed] [Google Scholar]
- 12.LeCun Y, Bengio Y, Hinton G. Deep learning. Nat 2015 5217553. 2015;521(7553):436–444. doi: 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 13.Géron A Hands-on Machine Learning with Scikit-Learn, Keras, and TensorFlow: Concepts, Tools, and Techniques to Build Intelligent Systems. O’Reilly Media; 2019. [Google Scholar]
- 14.McKinney W Data Structures for Statistical Computing in Python. Proc 9th Python Sci Conf. Published online 2010:56–61. doi: 10.25080/MAJORA-92BF1922-00A [DOI] [Google Scholar]
- 15.pandas development team T. pandas-dev/pandas: Pandas. Published online February 2020. doi: 10.5281/zenodo.3509134 [DOI] [Google Scholar]
- 16.Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res. 2011;12:2825–2830. [Google Scholar]
- 17.Chollet F, others. Keras. Published online 2015. https://github.com/fchollet/keras [Google Scholar]
- 18.Nudel J, Bishara AM, de Geus SWL, et al. Development and validation of machine learning models to predict gastrointestinal leak and venous thromboembolism after weight loss surgery: an analysis of the MBSAQIP database. Surg Endosc. Published online 2020. doi: 10.1007/s00464-020-07378-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the Areas under Two or More Correlated Receiver Operating Characteristic Curves: A Nonparametric Approach. Biometrics. 1988;44(3):837. doi: 10.2307/2531595 [DOI] [PubMed] [Google Scholar]
- 20.Lundberg SM, Allen PG, Lee S-I. A Unified Approach to Interpreting Model Predictions. Accessed October 21, 2021. https://github.com/slundberg/shap
- 21.T S, M L, ML T, MJ L, A H, JW M. A simple web-based risk calculator (www.anastomoticleak.com) is superior to the surgeon’s estimate of anastomotic leak after colon cancer resection. Tech Coloproctol. 2017;21(1):35–41. doi: 10.1007/S10151-016-1567-7 [DOI] [PubMed] [Google Scholar]
- 22.Rencuzogullari A, Benlice C, Valente M, Abbas MA, Remzi FH, Gorgun E. Predictors of anastomotic leak in elderly patients after colectomy: nomogram-based assessment from the American College of Surgeons National Surgical Quality Program Procedure-Targeted Cohort. Dis Colon Rectum. 2017;60(5):527–536. doi: 10.1097/DCR.0000000000000789 [DOI] [PubMed] [Google Scholar]
- 23.Rojas-Machado SA, Romero-Simó M, Arroyo A, Rojas-Machado A, López J, Calpena R. Prediction of anastomotic leak in colorectal cancer surgery based on a new prognostic index PROCOLE (prognostic colorectal leakage) developed from the meta-analysis of observational studies of risk factors. Int J Color Dis 2015 312. 2015;31(2):197–210. doi: 10.1007/S00384-015-2422-4 [DOI] [PubMed] [Google Scholar]
- 24.K M, D F, E V, et al. External Validation and Optimization of the French Association of Hepatopancreatobiliary Surgery and Transplantation’s Score to Predict Severe Postoperative Biliary Leakage after Open or Laparoscopic Liver Resection. J Am Coll Surg. 2018;226(6):1137–1146. doi: 10.1016/J.JAMCOLLSURG.2018.03.024 [DOI] [PubMed] [Google Scholar]
- 25.Yokoo H, Miyata H, Konno H, et al. Models predicting the risks of six life-threatening morbidities and bile leakage in 14,970 hepatectomy patients registered in the National Clinical Database of Japan. Medicine (Baltimore). 2016;95(49):e5466. doi: 10.1097/{MD}.0000000000005466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shinde RS, Acharya R, Chaudhari VA, et al. External validation and comparison of the original, alternative and updated-alternative fistula risk scores for the prediction of postoperative pancreatic fistula after pancreatoduodenectomy. Pancreatology. 2020;20(4):751–756. doi: 10.1016/j.pan.2020.04.006 [DOI] [PubMed] [Google Scholar]
- 27.Lao M, Zhang X, Guo C, et al. External validation of alternative fistula risk score (a-{FRS}) for predicting pancreatic fistula after pancreatoduodenectomy. {HPB} Off J Int Hepato Pancreato Biliary Assoc. 2020;22(1):58–66. doi: 10.1016/j.hpb.2019.05.007 [DOI] [PubMed] [Google Scholar]
- 28.Huang X-T, Huang C-S, Liu C, et al. Development and validation of a new nomogram for predicting clinically relevant postoperative pancreatic fistula after pancreatoduodenectomy. World J Surg. 2021;45(1):261–269. doi: 10.1007/s00268-020-05773-y [DOI] [PubMed] [Google Scholar]
- 29.Mungroop TH, van Rijssen LB, van Klaveren D, et al. Alternative Fistula Risk Score for Pancreatoduodenectomy (a-{FRS}): Design and International External Validation. Ann Surg. 2019;269(5):937–943. doi: 10.1097/{SLA}.0000000000002620 [DOI] [PubMed] [Google Scholar]
- 30.Tabchouri N, Bouquot M, Hermand H, et al. A novel pancreatic fistula risk score including preoperative radiation therapy in pancreatic cancer patients. J Gastrointest Surg. 2021;25(4):991–1000. doi: 10.1007/s11605-020-04600-y [DOI] [PubMed] [Google Scholar]
- 31.Han IW, Cho K, Ryu Y, et al. Risk prediction platform for pancreatic fistula after pancreatoduodenectomy using artificial intelligence. World J Gastroenterol. 2020;26(30):4453–4464. doi: 10.3748/wjg.v26.i30.4453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birkmeyer JD, Finks JF, O’Reilly A, et al. Surgical skill and complication rates after bariatric surgery. N Engl J Med. 2013;369(15):1434–1442. doi: 10.1056/{NEJMsa1300625} [DOI] [PubMed] [Google Scholar]
- 33.CP S, OA V, AM C, JD B, JB D. Video Ratings of Surgical Skill and Late Outcomes of Bariatric Surgery. JAMA Surg. 2016;151(6). doi: 10.1001/JAMASURG.2016.0428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.AB C, S L, JH N, Y L, AJ H. Machine learning analyses of automated performance metrics during granular sub-stitch phases predict surgeon experience. Surgery. 2021;169(5):1245–1249. doi: 10.1016/J.SURG.2020.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


