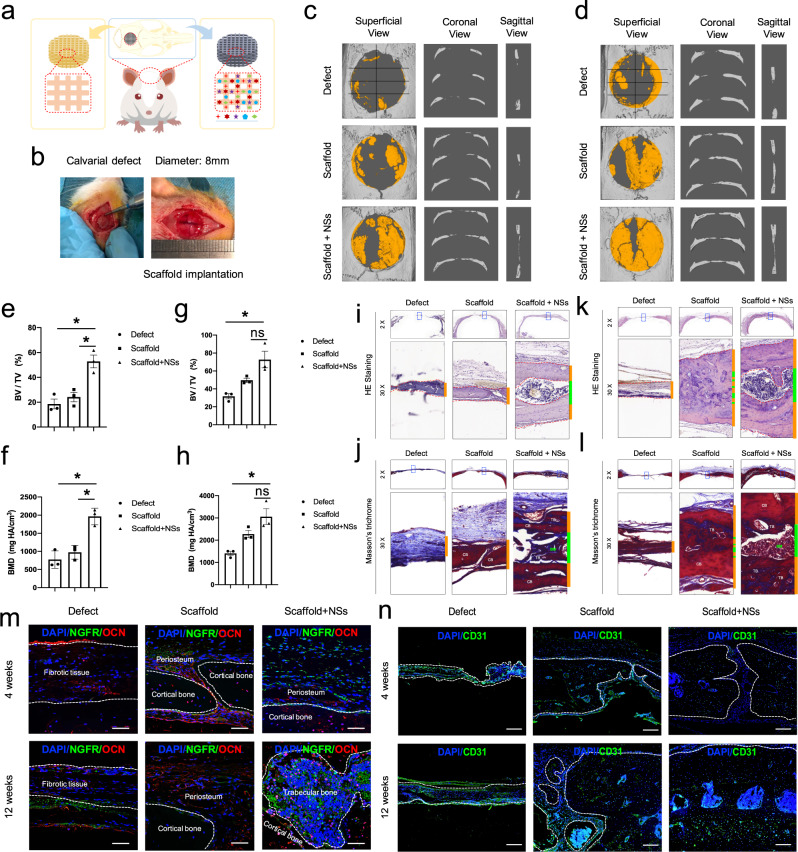
Fig. 2. Transplantation of the 3D printed hydrogel scaffold with NSs promotes rapid regeneration of critical-sized calvarial defect.
a, b Illustration of the implantation of NSs-loaded 3D printing gel scaffold before and after calvarial defect surgery. c, d μ-CT evaluation of bone regeneration in Defect control (Defect), 3D printed scaffold (Scaffold), 3D printed scaffold containing NSs (Scaffold + NSs) groups at 4 weeks and 3 12 weeks post-surgery of calvarial defect (the yellow area indicates new bone formation), respectively. n = 3 rats for per group and per time point. e Bone volume fraction (BV/TV) (Exact p value calculated with one-way ANOVA Tukey’s multiple comparisons test: *p = 0.0218, *p = 0.0391); and f Bone mass density (BMD) (Exact p value calculated with one-way ANOVA Tukey’s multiple comparisons test: *p = 0.0113, *p = 0.0214) of regenerated tissues in defect area at 4 weeks after calvarial defect. g Bone volume fraction (BV/TV) (Exact p value calculated with one-way ANOVA Tukey’s multiple comparisons test: **p = 0.0076, p = 0.0532); and h Bone mass density (BMD) (Exact p value calculated with one-way ANOVA Tukey’s multiple comparisons test: **p = 0.0045, p = 0.0582) of regenerated tissues in defect area 12 weeks after calvarial defect; n = 3 biologically independent rats. i, j HE staining and Masson’ Trichromic staining of paraffin sections in Defect, 3D Scaffold, Scaffold + NSs groups at 4 weeks after calvarial defect (bar = 500 μm at low magnification and bar = 50 μm at high magnification). k, l HE staining and Masson’ Trichromic staining of paraffin sections in Defect, 3D Scaffold, Scaffold + NSs groups at 12 weeks after calvarial defect (bar = 500 μm at low magnification and bar = 50μm at high magnification). m Co-immunofluorescent staining of NGFR and OCN expression in Defect, 3D Scaffold, Scaffold + NSs groups at 4 and 12 weeks after calvarial defect (bar = 50 μm), respectively. n Immunofluorescent staining of CD31 expression in Defect, 3D Scaffold, Scaffold + NSs groups at 4 and 12 weeks after calvarial defect (bar = 200 μm), respectively. The white dashed line indicates the border between the newly formed bone-like tissues. All data represent mean ± SD, and source data are provided as a Source Data file. At least three times of experiments were repeated independently.