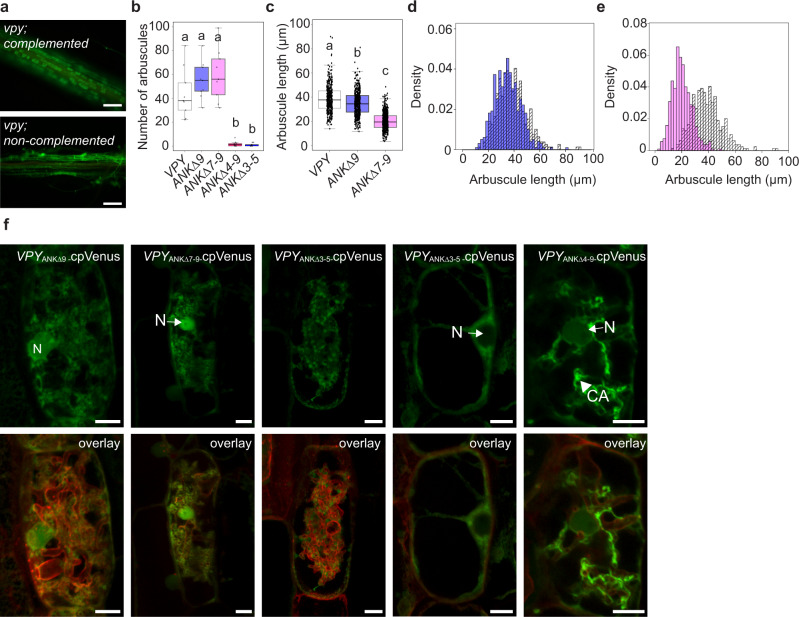
Fig. 2. Broadscale ankyrin deletion analysis.
a–e vpy-4 was transformed with VPYpro:VPY-cpVenus or ankyrin deletion constructs, colonized with Diversispora epigeae, and harvested 4 weeks post planting. Roots were stained with wheat germ agglutinin conjugated to AlexaFluor 488 (WGA-AlexaFluor 488) to visualize the fungus. a Representative epifluorescence images of vpy-4 WGA-AlexaFluor 488 stained roots complemented with VPYpro:VPY-cpVenus, or not complemented, as seen for vpy-4 transformed with VPYpro:VPYANKΔ3-5-cpVenus. Scale bars, 100 μm. b Number of arbuscules per infection unit in vpy-4 transformed with ankyrin deletion constructs. Different letters indicate statistically significant differences using one-way ANOVA, p < 0.05, n = 9 infection units, sampled from three biological replicates with three infection units per biological replicate. c Arbuscule length of vpy-4 transformed with VPYpro:VPY-cpVenus (n = 396 arbuscules), VPYpro:VPYANKΔ9-cpVenus (n = 496 arbuscules), or VPYpro:VPYANKΔ7-9-cpVenus (n = 536 arbuscules). Different letters signify statistically significant differences from one-way ANOVA, sampled from three biological replicates with three infection units per replicate. In (b)–(d), lines in boxplots represent the median value, box limits represent the upper and lower quartiles, whiskers represent 1.5 times the interquartile range. d Overlapping histogram of arbuscule length between VPYpro:VPYANKΔ9-cpVenus (purple bars) and VPYpro:VPY-cpVenus (striped bars). Arbuscule length is plotted on the x-axis, and probability densities are plotted on the y-axis to normalize sample size. e Overlapping histogram of arbuscule length between VPYpro:VPYANKΔ7-9-cpVenus (pink bars) and VPYpro:VPY-cpVenus (striped bars). Arbuscule length is plotted on the x-axis, and probability densities are plotted on the y-axis to normalize sample size. f Subcellular localization of VPY ankyrin deletions. WT roots expressing vectors containing VPYpro:VPYANKΔ9-cpVenus, VPYpro:VPYANKΔ7-9-cpVenus, VPYpro:VPYANKΔ3-5-cpVenus, or VPYpro:VPYANKΔ4-9-cpVenus and PM and PAM marker BCP1pro:PT1-mCherry were colonized with R. irregularis to determine the subcellular distribution of the truncated fusion proteins in arbuscule-containing cells. Overlay images display both cpVenus and mCherry channels. Samples are representative images of at least two independent experiments, with three root systems and ten root pieces imaged per experiment, n = 47 (VPYpro:VPYANKΔ9-cpVenus), n = 19 (VPYpro:VPYANKΔ7-9-cpVenus), n = 44 (VPYpro:VPYANKΔ4-9-cpVenus), or n = 22 (VPYpro:VPYANKΔ3-5-cpVenus) arbuscule-containing cells. Fluorescence images are projections of 10 optical sections on the z-axis taken at 0.5 µm intervals. Scale bars, 10 µm. N, nucleus (arrow); CA, cytoplasmic aggregate (arrowhead).