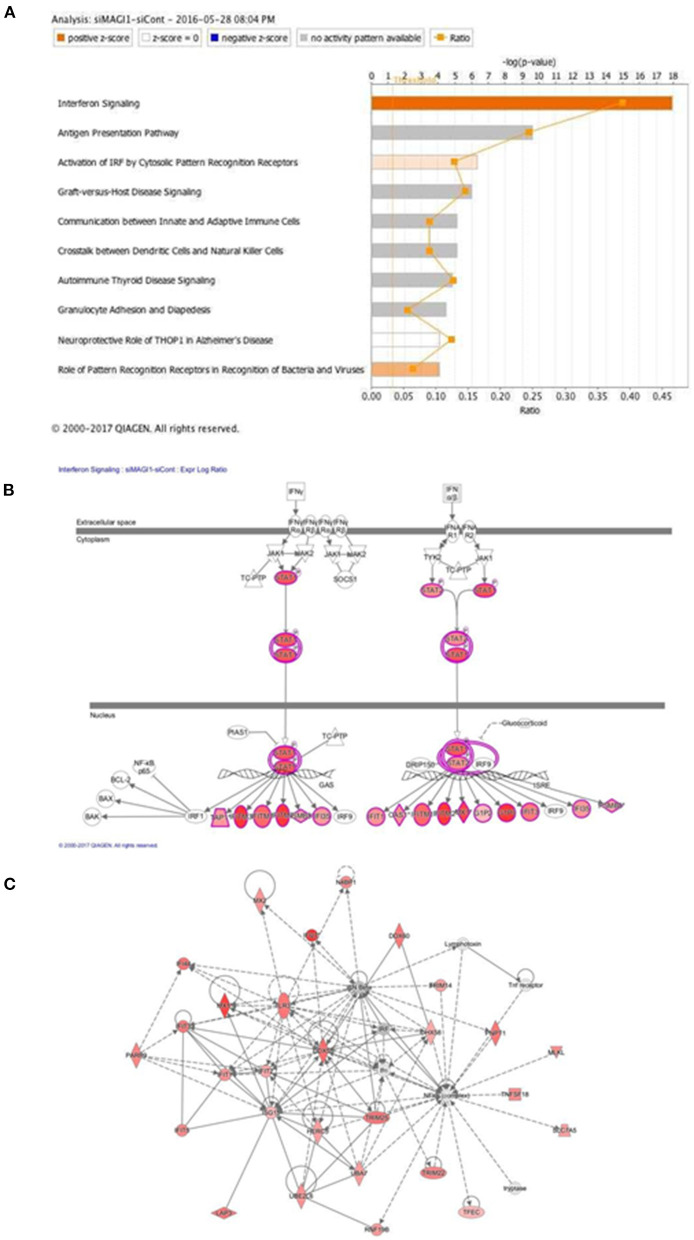
Figure 4.
IFN signaling is the top-rank enriched IPA canonical pathway. (A) IPA-based identification of canonical pathways (–log[P] >5) associated with differentially expressed genes in HUVECs transfected with siMAGI1 vs. siCont. The bar chart indicates the –log(P) value of the significance of enrichment of each category. (B) Induction of IFN signaling in MAGI1-depleted ECs. Pathway models of IFN signaling in ECs based on data in the Ingenuity Knowledge Base are shown. Expression of genes in red is higher in siMAGI1-treated ECs compared to that in siCont-treated ECs. (C) Gene enrichment analyses (right-tailed Fisher exact test) according to the IPA downstream effects analysis identified Antimicrobial Response, Inflammatory Response, and Infectious Diseases as the highest scoring IPA interaction networks. Green and red symbols denote genes with lower and higher expression, respectively, in siMAGI1-treated ECs compared to that in siCont-treated ECs. The arrows with solid lines indicate direct (usually physical) interactions between two molecules in the direction of the arrow, Whereas, arrows with dashed lines denote indirect interactions (e.g., molecule/gene X affects molecule/gene Y). The abbreviations shown are defined in Supplementary Table 6.