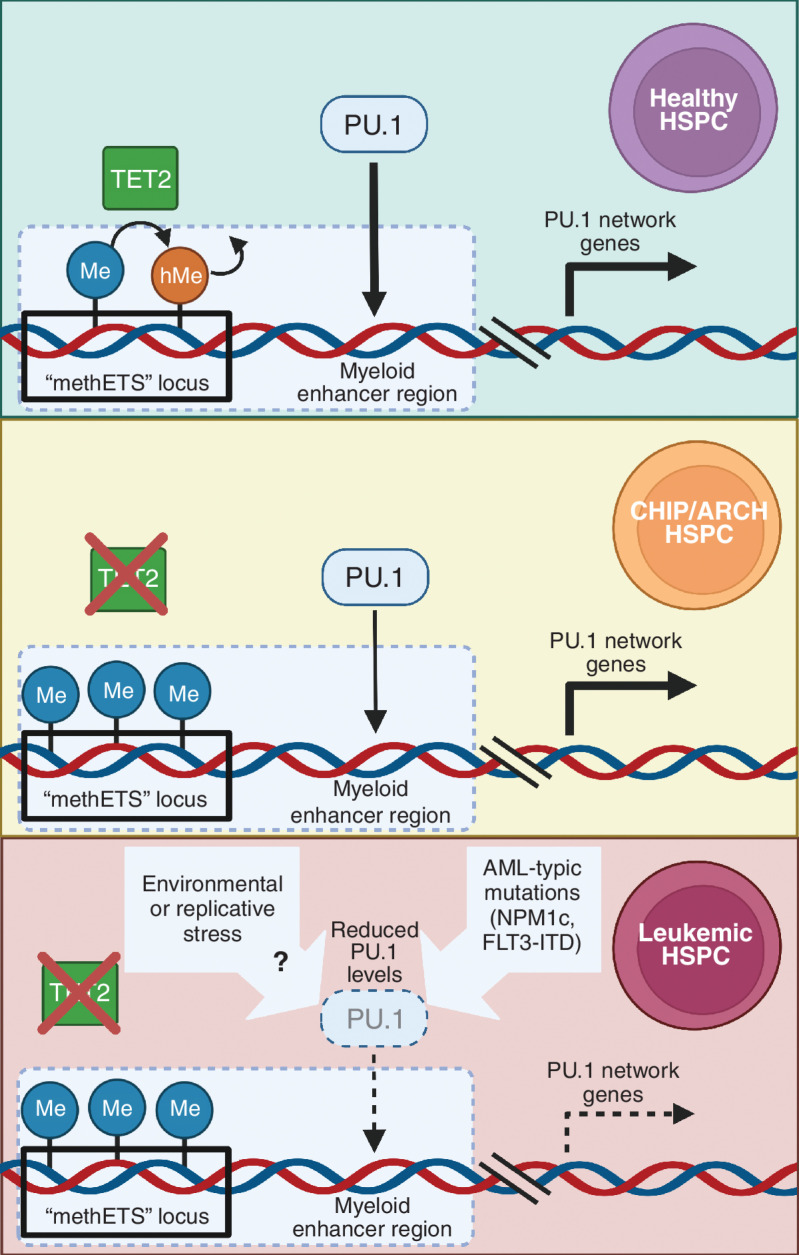
Figure 1.
In healthy hematopoietic stem and progenitor cells (HSPC), TET2 promotes the first step in cytosine demethlation, the conversion of methylcytosine (Me; blue lollipop icons) to hydroxymethylcytosine (hMe; orange lollipop icons) residues at the enhancer regions of myeloid target genes. This allows appropriate PU.1 binding and subsequent transactivation of myeloid maturation genes. In the context of TET2 deficiency associated with clonal hematopoiesis of indeterminate potential/age-related clonal hematopoiesis (CHIP/ARCH) loss of TET2 activity in HSPC results in hypermethylation of specific ETS binding sites located in many PU.1 target gene enhancers (termed by the authors as methylation-sensitive ETS, or methETS, loci; depicted by black box outline) which in turn may reduce the capacity for PU.1 to transactivate gene expression at these targets (depicted by reduced thickenss of the PU.1 arrow). AML-typic mutations and potentially other aging-related environmental factors that reduce PU.1 expression can act in concert with hypermethylation of methETS loci to further abolish PU.1 binding at these regions (depicted by broken lines around PU.1 and arrow). Resultant dysregulation of the PU.1 network ultimately results in leukemic transformation of HSPC.