Abstract
The (E)-β-farnesene (EβF) is one of the most important secondary metabolites in some plants and provides indirect defense against aphids. However, the direct effect of EβF against pests is still unclear. In this study, various concentrations of EβF (0.16, 0.8, and 4 g/kg) were provided in an artificial diet to determine the direct effects of EβF on Spodoptera exigua. The results showed that an artificial diet containing 4 g/kg of EβF reduced the final survival of the S. exigua larvae and per female fecundity of adults significantly when compared with CK and SC controls (p < 0.05), then ultimately it also significantly affected the intrinsic rate of increase (p < 0.05). Furthermore, the results of the EβF bioassay in an artificial diet also indicated that the proliferation of the S. exigua population was inhibited by the ingestion of EβF in a dose-dependent manner. Combined differential RNA-seq data and RT-qPCR analysis, it was found that four key genes involved in juvenile hormone degradation significantly upregulated in S. exigua larvae treated by EβF at a dose of 0.8 and 4 g/kg when compared with two controls (p < 0.05). This indicated that EβF could disturb the normal function of juvenile hormones and reduce the survival rate of S. exigua larvae. Additionally, two key genes that regulate per fecundity of S. exigua females, including SeVg and SeVgR, were significantly downregulated in adult females (p < 0.05) when they were treated with 0.8 and 4 g/kg of EβF at the larval stage, relative to the expression of these genes after treatment with controls. These findings suggested that EβF first disturbed the normal function of juvenile hormone by upregulating key degradation genes, and then inhibited the expression of SeVg/SeVgR genes and proteins, thus reducing the population size of S. exigua by increasing larval mortality and inhibiting per female fecundity.
Keywords: Spodoptera exigua, (E)-β-farnesene, RNA-seq, SeVg and SeVgR, reproduction and oviposition
Introduction
Spodoptera exigua is a major polyphagous pest that affects crops, such as alfalfa, soybean, peanut, onion, corn, scallion, and cotton, leading to huge economic losses (Saeed et al., 2010; Zhang et al., 2011; Hafeez et al., 2020). S. exigua is mainly controlled by spraying chemical pesticides (Saeed et al., 2019; Huang et al., 2021). However, the development of insecticide resistance (one of the “3R” major concerns) occurs due to the long-term and improper use of pesticides on agricultural products (Zhen and Gao, 2016). To prevent the target insects from developing resistance to insecticides, novel bioactive molecules with high efficiency but low toxicity to non-target organisms are required in agricultural production (Zhen and Gao, 2016).
Plants have co-evolved with insects and produce various secondary metabolites to defend against pests (Hu et al., 2021). The secondary metabolites from plants can be divided into four categories: Terpenoids, phenols, alkaloids, and sulfur-containing compounds. These metabolites exhibit toxicity, antifeedant activity, and antibiosis effects on pests (Adeyemi, 2010; War et al., 2012). Among these phytochemicals, terpenoids are the most structurally diverse (Nagegowda and Gupta, 2020). Terpenoids are synthesized from the five-carbon isopentenyl diphosphate (IPP) and isomer dimethylallyl diphosphate (DMAPP) (Sun et al., 2017; Michael et al., 2020; Zhou and Pichersky, 2020). With the advancement in molecular biology, many key enzymes that are required for the synthesis of terpenes [e.g., terpene synthases (TPSs)] have been identified in Arabidopsis thaliana (Chen et al., 2004).
Recently, many TPS coding genes were found in gramineous crops, such as TPS10 in corn, TPS46, and TPS23 in rice. These genes play a key role in indirect defense against insect pests (Yuan et al., 2008; Capra et al., 2015; Sun et al., 2017). Among the products of these three TPS genes, the products of TPS10 and TPS46 have very similar amino acid sequences (Sun et al., 2017). Interestingly, both TPS10 and TPS46 synthesize the same volatile substance (E)-β-farnesene (EβF) (Schnee et al., 2006; Yuan et al., 2008; Sun et al., 2017).
The compound EβF plays an important role in crop pest control, especially in controlling aphids (Beale et al., 2006; Cui et al., 2012). Overexpression of tps10 in A. thaliana not only repelled aphids but also attracted aphid parasitoids, whereas the overexpression of tps46 in rice also repelled bird cherry-oat aphid (Beale et al., 2006; Sun et al., 2017). The latest research showed that EβF had presented a magic attractive effect on the larvae and adults of the aphid predator hoverfly Eupeodes corollae (Wang et al., 2022). Additionally, EβF was also used to attract the natural enemies of cabbage aphids to control these pests in Chinese cabbage fields (Cui et al., 2012; Su et al., 2015). With the advancement in molecular biology, the TPSs of EβF have also been identified in many other plants, such as Matricaria recutita (L.), Douglas-fir, and Mentha piperita (Huber et al., 2005; Yu et al., 2013; Su et al., 2015).
Terpenes from many plants have been reported to be toxic to pests. Four monoterpenes, including β-phellandrene, α-pinene, p-paracymene, and (+)-2-mainly carene, produced in greenhouse tomato flowers, showed direct toxicity to insects (Morse et al., 2012). Previous studies also showed that EβF, as one of the important products of TPS46, has a direct lethal effect on many other phytophagous insects (Cui et al., 2012). Therefore, EβF and the TPSs associated with it might be applied as substitutes for chemical pesticides to protect plants. In our previous study, EβF showed a strong lethal effect on the larvae of the Chilo suppressalis, and consequently, C. suppressalis failed to complete its life cycle (Yang et al., 2022). However, S. exigua could complete its life cycle when treated with the same concentration of EβF as used for controlling C. suppressalis. This indicated that there was a difference in the response of S. exigua and C. suppressalis to EβF. Therefore, to determine the mechanism by which EβF affects the life cycle of insects, especially that of adults. We used differential RNA-seq, Western blotting, and bioinformatics techniques to systematically analyze the life table data and the expression of key genes in S. exigua after treatment with different sublethal doses of EβF. Therefore, the physiological and molecular responses of S. exigua after being treated with EβF were elucidated, which might provide a basis for determining the natural compounds to control S. exigua in crops.
Materials and methods
Rearing of Spodoptera exigua
The initial colony of S. exigua was provided by the Institute of Plant Protection, Jiangsu Academy of Agricultural Sciences, and the larvae were raised on an artificial diet following an established method (Ren et al., 2013) in the laboratory without exposure to pesticides at 27 ± 1°C, relative humidity of 65 ± 5% under a 14:10 light/dark cycle. The adults of S. exigua were provided with 10% honey solution as food in a previous study (Zhao et al., 2018).
Preparing different concentrations of (E)-β-farnesene for Spodoptera exigua treatments
The EβF was added to the artificial diet of the larvae (Yang et al., 2022). Based on our previous study (Sun et al., 2017), the five concentration gradients of EβF (18794–84–8, Sigma-Aldrich), including 0.032, 0.16, 0.8, 4, and 20 g/kg (EβF/artificial diet, standard compound of EβF/uncured liquid feed), were set for initial testing. In the pre-experiment phase, treatment of S. exigua larvae with 0.032 g/kg of EβF showed no noticeable effect, while treatment with 20 g/kg of EβF killed all the larvae. Meanwhile, the standard EβF purchased from Sigma-Aldrich was very expensive, and the use of 20 g/kg of EβF/artificial diet required huge consumption. Additionally, the mortality of S. exigua larvae treated with 4 g/kg of EβF/artificial diet was close to 50%, which was equivalent to the lethal medium concentration (LC50). Therefore, 4 g/kg of EβF/artificial diet was set as the maximum treatment dose by considering the cost. Next, three concentration gradients of EβF were selected (0.16, 0.8, and 4 g/kg), as well as ethanol and water treatments were used as solvent control (SC) and blank control (CK), respectively. The newly hatched first instar larvae of S. exigua were exposed to five treatments, including 0.16, 0.8, 4 g/kg, SC, and CK, and their entire life cycle was monitored for recording specific effects during different stages of development. In the larval stage, the mixed artificial diet was constantly changed to maintain the dosage stability of EβF on the larvae. The S. exigua larvae were placed in a small perforated plastic box (10 cm × 6 cm × 5 cm) and fed with an artificial diet. Each treatment had five replicates, and each replicate had 200 larvae, which were maintained in 40 small plastic boxes (five larvae in each box).
Sample collection and construction of the life table
Following previously described methods (Sun et al., 2016; Zhao et al., 2018), the life table of S. exigua was constructed and included data on the mortality of the larvae at different instars, per female fecundity, the hatching rate, the intrinsic rate of increase, and other key parameters (Table 1). The parameters were recorded for each treatment from multiple insects maintained in separate boxes. The data collected from multiple insects were summarized to construct the life table.
TABLE 1.
The key life-history parameters of S. exigua larvae under different doses of EβF.
| Key life history parameters of S. exigua | Different treatments | ||||
|
|
|||||
| Controls | Different doses of Eβ F | ||||
|
|
|
||||
| CK | SC | 0.16 g/kg | 0.8 g/kg | 4 g/kg | |
| Total mortality of larvae at 1st and 2nd instars (%) | 4.38 ± 0.87c | 3.33 ± 1.36c | 5.00 ± 1.71c | 11.67 ± 4.38b | 28.33 ± 6.18a |
| Larvae mortality of 3rd instar (%) | 1.46 ± 0.93a | 1.04 ± 0.74a | 1.67 ± 1.58a | 2.08 ± 1.65a | 2.71 ± 0.94a |
| Larvae mortality of 4th instar (%) | 1.88 ± 1.36a | 2.29 ± 1.36a | 1.87 ± 0.87a | 1.46 ± 1.19a | 2.50 ± 1.19a |
| Ultimate larvae mortality (%) | 14.17 ± 1.58c | 17.50 ± 2.00c | 19.17 ± 2.72c | 36.67 ± 5.02b | 54.75 ± 7.10a |
| Larval stage (d) | 12.64 ± 1.64a | 12.78 ± 1.94a | 13.13 ± 1.92a | 13.55 ± 2.52a | 14.12 ± 3.08a |
| The whole life-span of S. exigua (d) | 31.19 ± 3.02a | 31.37 ± 3.04a | 31.65 ± 3.37a | 32.79 ± 4.30a | 34.43 ± 6.04a |
| Adult emergence rate (%) | 78.25 ± 9.98a | 79.11 ± 7.93a | 74.66 ± 8.71a | 72.70 ± 8.77a | 70.52 ± 8.15a |
| Per female fecundity | 512.40 ± 57.67a | 504.80 ± 45.86a | 464.20 ± 62.01a | 370.00 ± 54.19b | 155.40 ± 32.11c |
| Hatching rate of eggs (%) | 68.47 ± 7.14a | 67.11 ± 6.92a | 66.60 ± 7.68a | 63.38 ± 6.79a | 57.98 ± 6.89a |
| Intrinsic rate of increase (R) | 0.154 ± 0.018a | 0.151 ± 0.012a | 0.144 ± 0.017a | 0.122 ± 0.019b | 0.079 ± 0.012c |
CK (blank control) and SC (solvent control) were set as controls. The concentrations of EβF treatments were set as 0.16, 0.8, and 4 g/kg; the units of g/kg in all treatment groups mean the mass ratio of EβF/artificial diet. Data are presented by mean ± standard error; different lower-case letters indicate a significant difference obtained by ANOVA, followed by Tukey’s HSD test (p < 0.05).
Adult S. exigua were placed in a lidless plastic box (20 cm × 20 cm × 20 cm) with the top covered with gauze to lay eggs in the dark. Before hatching, the eggs were transferred to sterile glass jars and administered an artificial diet. They were incubated under normal conditions (27 ± 1°C; 65 ± 5% RH; 14:10 light/dark cycle). To determine the hatching rate, five groups of egg clothes were randomly selected to count the number of eggs in a clutch under a microscope, and then the number of hatched larvae was counted to evaluate the hatching rate. To determine per female fecundity, 10 pairs of S. exigua adults were selected from each treatment group, and the experiment was repeated five times. The per female fecundity was evaluated by calculating the average number of eggs laid for each pair of male and female adults.
During the experiment, S. exigua was sampled twice; the initial sampling was performed at the beginning of the third instar of S. exigua, and the final sampling was performed when the adult females were 2 days old. The initial sampling was performed at the beginning of the third instar because the first and second instar EβF-treated larvae had high mortality, but the third and fourth instar larvae had almost no deaths. We sampled 50 third instar larvae of S. exigua for each treatment, and five repeats were performed. Half of the samples were used for performing differential RNA-seq to detect the changes in the key genes in the surviving S. exigua larvae, while the other half of the samples were used for performing real-time quantitative PCR (RT-qPCR).
The second sampling was performed when the adult females were 2 days old to measure the gene and protein expressions of SeVg and SeVgR. Additionally, the fat body and tissue samples of the ovary from these adult females treated with different doses of EβF were dissected and collected. Preliminary experiments showed that EβF treatment strongly inhibited per female fecundity of S. exigua. Our previous study also showed that SeVg and SeVgR are the key proteins for evaluating the reproductive potential of adult females and their expression in female S. exigua can reach the maximum level within 2 days (Sun et al., 2016; Zhao et al., 2018). Therefore, 10 2-day-old female adults were taken from each treatment and repeated the experiment three times. Thus, a total of 30 females were used to detect the expression of SeVg and SeVgR proteins. Additionally, two 2-day-old adult females were taken for each treatment, repeated five times, and a total of 20 females were used to detect the expression of SeVg and SeVgR genes by performing RT-qPCR.
RNA isolation, library preparation, and PacBio sequencing
Total RNA has extracted from S. exigua larvae at the beginning of the third instar using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) strictly as directed by the manufacturer. The integrity of the RNA was determined with the Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, California, United States). Total RNA samples with RIN value ≥ 8 were used for constructing the cDNA libraries in PacBio sequencing. Using the Clontech SMARTer PCR cDNA Synthesis Kit (Takara Biotechnology, Dalian, China), 4 μg RNA is synthesized to cDNA and subsequently amplified to generate double-stranded cDNA. The cDNA was then the size selected for < 4 kb and > 4 kb fractions using the BluePippin Size Selection System (Sage Science, Beverly, MA, United States). Each SMRTbell library was constructed using 1 μg size-selected cDNA with the Pacific Biosciences SMRTbell template prep kit. The binding of SMRT bell templates to polymerases was conducted using the Sequel II Binding Kit, and then primer annealing was performed. Sequencing was carried out on the Pacific Bioscience Sequel II platform.
For short-read sequencing, magnetic beads with Oligo (dT) were used to enrich eukaryotic mRNAs for constructing the sequencing library. Next, a fragmentation buffer was added to break the mRNAs into short fragments. Then, the RNA-seq libraries were constructed using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (NEB, United States), following the manufacturer’s instructions. The different libraries were pooled for sequencing on the Illumina HiSeq platform. Sequencing of all samples was conducted by Nanjing Genepioneer Biotechnologies Inc. (Nanjing, China).
Unigene assembly and annotation
The standard protocol of ISO-seq (SMRT Analysis 2.3) was followed to process the raw PacBio full-length ISO-seq data. The reads of inserts (ROIs) were obtained from the circular consensus sequences (CCS). After searching for 5’ and 3’ adaptors and poly(A) signals, full-length, and non-full-length cDNA reads were defined, and chimeric reads were removed, including the sequencing primers. The redundant sequences were moved using CD-HIT-EST to obtain non-redundant (NR) high-quality transcripts (Fu et al., 2012). The sequences of these NR transcripts were defined as unigenes.
To determine the open reading frames (ORFs) present in the transcripts, we used TransDecoder v2.011 to confirm the putative coding sequences (CDSs). The predicted CDSs were searched and confirmed by BLASTX (E-value ≤ 1e-5) against three protein databases [NR, SwissProt, and Kyoto Encyclopedia of Genes and Genomes (KEGG)]. The transcripts containing complete ORFs, 5’-untranslated regions (UTRs), and 3’-UTRs were regarded as FL transcripts. For further studying the juvenile hormone-related genes, we selected the genes related to juvenile hormone based on the information on the annotation of unigenes.
Analysis of differentially expressed genes
Fastp v0.20.1 (Chen et al., 2018) was used to remove the adapters and low-quality sequences in the raw data. The short reads of each sample were aligned to the unigenes using Bowtie2 (Langmead et al., 2009). The read counts of the unigenes were obtained using htseq-count (Anders et al., 2015). The FPKM (Fragments Per Kilobase of transcript per Million mapped reads) of the unigenes in all samples were calculated as the expression level. RSEM was used to calculate the value of the FPKM (Li and Dewey, 2011). DESeq2 was used for differential expression analysis, and the unigenes with a cut-off of | log2 ratio| ≥ 1 and q < 0.05 were selected for significant differential expression (Love et al., 2014).
The FPKM values of the differentially expressed juvenile hormone-related genes were converted to log2 (FPKM) values. Then, the log2 (FPKM) values were used to plot the heatmap by ComplexHeatmap (Gu et al., 2016).
RT-qPCR analysis
Total RNA of the S. exigua samples exposed to different treatments (CK, SC, and different doses of EβF) was extracted using the TRIzol reagent (Invitrogen, Carlsbad, CA, United States) strictly as directed by the manufacturer. An MMLV Reverse Transcriptase (Promega, Madison, WI, United States) treated with ribonuclease H (Takara, Tokyo, Japan) was used to synthesize the cDNA library, and spectrophotometry was performed to quantify the library. The primers used for RT-qPCR (SYBR Green I) were designed based on the genes that were found to be related to the juvenile hormone from the RNA-seq data (Table 2). Also, the house-keeping gene of S. exigua, β-actin was used as an endogenous reference for data normalization (Zhao et al., 2018). The primers of the house-keeping gene and the other 10 random gene sequences that were used to verify the results of differential RNA-seq are listed in Table 2. And the details of these 10 selected genes are listed in Supplementary Table 1.
TABLE 2.
Sequences of RT-qPCR primers used in this study.
| Gene | Upstream primer(5′-3′) | Downstream primer (5′-3′) | Tm (°C) | Product size (bp) |
| SeVg | 5′-GCATACCAGCCAACTACCAAAT-3′ | 5′-TGCACCTGACACTGTCTACCCT-3′ | 60°C | 149 |
| SeVgR | 5′-GAAGGGAGGGAAGTGTCCTGAG-3′ | 5′-TGATGGTGAAAGAAACGCTGTG-3′ | 60°C | 104 |
| Actin | 5′-CCAGCCTTCCTTCTTGGGTAT-3′ | 5′-AGGTCCTTACGGATGTCAACG-3′ | 60°C | 94 |
| Unigene026066 | 5′-AGTTTGATGAAGCGATGA-3′ | 5′-TTAGTCTTGAAATGAGGGA-3′ | 50°C | 199 |
| Unigene025818 | 5′-ATCCCAACCACAGCACCAG-3′ | 5′-AATGCCCTCCATCATCCAG-3′ | 58°C | 127 |
| Unigene025841 | 5′-GCAATGGGTTTGGCTACT-3′ | 5′-TTTGTTGGCTGGGTCTGT-3′ | 54°C | 232 |
| Unigene024849 | 5′-TATGGGTTTATGTGCTTAGA-3′ | 5′-GTAGAACTGATGTGGCTCC-3′ | 50°C | 163 |
| Unigene022385 | 5′-GTATTAGCGAGGGAAACA-3′ | 5′-ACCAATCTGAGCAGCACT-3′ | 53°C | 201 |
| Unigene029506 | 5′-TGATGATGCTGGCTGAGAT-3′ | 5′-GAGGCTGGTGAAACAACTG-3′ | 60°C | 286 |
| Unigene029881 | 5′-CCCCGTCAGCAAGGTGG-3′ | 5′-CGTTGGCGTTGAATCCG-3′ | 60°C | 214 |
| Unigene029782 | 5′-GCCCACCCCAAATACGA-3′ | 5′-GTCAGCCTCCACCAGCG-3′ | 57°C | 123 |
| Unigene024091 | 5′-GTTGATGGGTGATTTGG-3′ | 5′-TACACTGGGTTGGTCGT-3′ | 50°C | 105 |
| Unigene024366 | 5′-GGAGTGTAAACAGAAGGGAGTC-3′ | 5′-GCTGGTCGCTGATGAAGA-3′ | 54°C | 162 |
The Primer 5.0 software was used to design all EST-specific primers. The reactions were performed using the SYBR Premix Ex Taq Kit (Takara, Tokyo, Japan) and were conducted on the Bio-Rad iCycler real-time quantitative RT-PCR detection system. The RT-qPCR was conducted using a 25 μL reaction mixture, which contained 0.5 μL of each primer (10 μM) (total 1.0 μL), 12.5 μL of 2 × SYBR Premix Ex Taq, 2.0 μL of sample cDNA (100 ng), 0.5 μL of ROX Reference DYE, and sterilized H2O 9.0 μL. And the RT-qPCR parameters were set as follows: 5 min at 94°C; (35 cycles) 10 s at 94°C, 20 s at 53–58°C, and 15 s at 75°C. The melting curve analysis and gel electrophoresis were also performed to confirm the RT-qPCR quality of these genes. The reactions of each treatment were replicated four times; non-template control reactions were performed in triplicate for each primer pair. The relative expression levels of each gene in S. exigua exposed to different treatments (expressed as the relative quantification (RQ) values) were calculated using the 2–ΔΔ Ct method (Livak and Schmittgen, 2001). Significant differences in the expression of the genes were determined by performing a one-way analysis of variance, followed by Duncan’s multiple comparison test.
The relative expression SeVg and SeVgR in Spodoptera exigua treated with different doses of (E)-β-farnesene
SeVg and SeVgR were cloned in a previous study (Zhao et al., 2018). The specimens were snap-frozen in liquid nitrogen and kept at –80°C until used for total RNA extraction and RT-qPCR. Each treatment group had five biological replicates, with two adult females per replicate. The primers used for performing RT-qPCR of the SeVg and SeVgR genes were based on a previous study (Zhao et al., 2018), and are listed in Table 2.
The relative expression of SeVg and SeVgR proteins in female adults treated with different doses of (E)-β-farnesene
The fat body and ovary tissue samples of more than 10 females were used to perform the Western blot analysis (Sun et al., 2016; Zhao et al., 2018). Total proteins were extracted using the Tissue Protein Extraction Reagent kit (Zoonbio Biotechnology Co., Ltd, Nanjing, China), and the concentrations of the proteins were determined by the bicinchoninic acid (BCA) method. The Western blot analysis was performed following the methods reported in a study by Song et al. (2012), with some modifications using the grayscale value of target protein/grayscale value of β-actin protein. Uniform spotted wells were used, the concentration of S. exigua protein in each well was equal (1 μg/μL), and 20 μL of the sample was loaded to ensure that each well contained about 20 μg. For each Western blot analysis, samples containing an equal amount of total proteins were resolved on the same gel. Protein samples were electrophoresed on 10% SDS-PAGE and electroblotted onto an NC membrane (Bio-Rad) running at 100 mA for 3 h with Tris/glycine buffer. The membranes were then blocked for 1 h at 37°C with 5% non-fat powdered milk in Tris-buffered saline containing 0.05% Tween 20 (TBS-T).
The primary antibody was specific and produced by the Zoonbio Biotechnology Co., Ltd, Nanjing, China. It is based on the SeVg and SeVgR gene and protein sequence. The 697 amino acids (aa) specific antibody sequence of SeVg was designed from 29 to 725 aa sequence of SeVg full-length protein (1,761 aa, accession number AOH73254.1 at the National Center for Biotechnology Information, NCBI), which contained 2,091 base pairs (bp) designing from 85 to 2,175 bp in the SeVg mRNA complete CDS (5,286 bp, accession number KT599434.1 in NCBI). The 256 aa specific antibody sequence of SeVgR was designed from 1,063 to 1,318 aa sequence of SeVg full-length protein (1,814 aa, accession number AOX13593.1 in NCBI), which contained 768 bp designed from 3,187 to 3,954 bp in the SeVgR mRNA complete CDS (5,445 bp, accession number KT899978.1 in NCBI). Then, the primary antibody was incubated overnight with the sample. The secondary antibody was a goat anti-rabbit antibody procured from the Boster Biological Technology (Catalog no. BA1054), which was incubated with the sample for 1–2 h.
Statistical analysis
Statistical analyses were conducted using the SAS v.9.0 software (SAS Institute, Cary, NJ, United States). The data were presented as the mean ± standard deviation and analyzed by performing a one-way analysis of variance (ANOVA), followed by Tukey’s honest significant difference (HSD) test. According to Greenberg et al. (2001); Sun et al. (2016), and Zhao et al. (2018), the relationships between the key life-history parameters of S. exigua and the relative expression of SeVg or SeVgR genes were evaluated by linear regression analyses.
Results
(E)-β-farnesene effects on Spodoptera exigua at different concentration treatments
S. exigua treated with EβF/artificial diet (including 0.16, 0.8, and 4 g/kg) showed lower food intake, higher developmental disorder, higher mortality of the first and second instar larvae, and lower per female fecundity than that treated with CK and SC controls. The effects on the key life-history parameters of S. exigua larvae treated with different lethal doses of EβF are shown in Table 1. Non-significant differences were observed in per female fecundity, hatching rate of eggs, and intrinsic rate of increase between S. exigua individuals treated with 0.16 g/kg of EβF and those in the two controls (p > 0.05; Table 1). Nevertheless, compared with the CK and SC controls, the S. exigua larvae treated with EβF at the concentrations of 0.8 g/kg and 4 g/kg had a significant increase in the mortality rate of first and second instar larvae (p < 0.05; Table 1). The mortality rate of first and second instar larvae treated with 0.8 g/kg EβF was 11.67%, and with 4 g/kg could reach 28.33%. No significant difference was seen in the mortality of the third and fourth instar larvae treated with EβF at the doses of 0.8 and 4 g/kg in comparison with two controls (p > 0.05; Table 1). Significant differences in ultimate mortality of S. exigua larvae treated with EβF at the concentration of 0.8 and 4 g/kg (36.67 and 54.75%, respectively) were observed (p < 0.05; Table 1). These results showed that larval mortality was mostly affected by EβF in a dose-dependent manner, especially at 4 g/kg (p < 0.05; Table 1).
Compared with CK and SC controls, there were no significant differences in life-span, adult emergence rate, and hatching rate of eggs between 0.8 and 4 g/kg EβF treatments (p > 0.05; Table 1). However, two parameters of population build-up of S. exigua, i.e., per female fecundity and intrinsic rate of increase, had significant differences after treatment with 0.8 and 4 g/kg of EβF (p < 0.05; Table 1). The EβF concentration of 4 g/kg had the strongest effect on per female fecundity and the intrinsic rate of increase (R) in S. exigua larvae (p < 0.05; Table 1). These results indicated that the effect of EβF on S. exigua was stronger at higher doses.
Significant differences in larval survival and adult oviposition of S. exigua were found after treatment with 0.8 g/kg EβF (p < 0.05; Table 1) compared to the control, and the differences were more prominent at 4 g/kg of EβF (p < 0.05; Table 1). Thus, 0.8 and 4 g/kg of EβF effectively inhibited the growth of the S. exigua population (p < 0.05; Table 1).
Functional annotation and differential expression analysis of unigenes
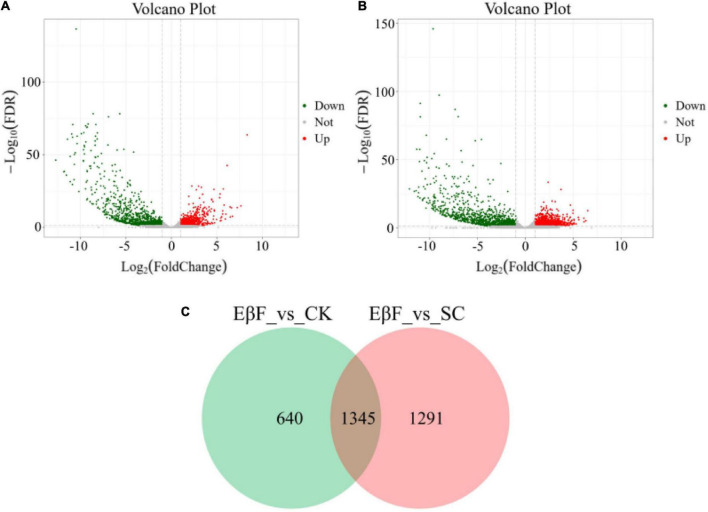
The Pacbio sequencing platform generated 34,339 unigenes with an N50 value of 4,625 bp. A total of 30,009 unigenes were annotated in at least one public database, such as NR, NT, COG, GO, KEGG, Swissport, and Interpro protein databases. Finally, 17,473 unigenes were found to be expressed in at least one sample. Differential gene expression analysis was conducted between the samples in the EβF treatments and controls (SC and CK) with fold change levels ≥ 2 with an FDR ≤ 0.05. It resulted in the identification of 1,985 different expressed genes between EβF treatment and CK (Figure 1A). Of these, 1,161 unigenes were downregulated, while 824 genes were upregulated under EβF treatments. Meanwhile, 2,636 differentially expressed genes (DEGs) were identified between EβF treatments and SC (Figure 1B). In these DEGs, 1,372 were downregulated DEGs, and 1,264 were upregulated DEGs. Among these DEGs, 1,345 unigenes were differentially expressed both in the 4 g/kg vs. CK and 4 g/kg vs. SC (Figure 1C and Supplementary Tables 2, 3).
FIGURE 1.
The mixed graphs showed the relative expression level of all differentially expressed unigenes in S. exigua larvae after EβF treatments at doses of 4 g/kg. (A) The volcano plot obtained from the DESeq2 analysis between 4 g/kg EβF treatments and CK (blank control) samples showing log2 (fold-change) and-Log10 FDR values. (B) The volcano plot obtained from DESeq2 analysis between 4 g/kg EβF treatments and SC (solvent control) samples showing log2 (foldchange) and-Log10 FDR values. (C) The numbers of DEGs among different treatments among CK, SC, and 4 g/kg EβF treatment. The concentrations of EβF treatments were set as 4 g/kg; the unit of EβF treatment as g/kg means the mass ratio of EβF/artificial diet. The value denotes the value of log2 (FPKM + 1).
Based on the annotation information, 48 juvenile hormone-related unigenes were identified. Eighteen juvenile hormone-related genes were differentially expressed between EβF treatment and CK. Additionally, 25 juvenile hormone-related genes were differentially expressed between the EβF treatment and the SC control (Figure 2). Among these differentially expressed juvenile hormone-related unigenes, 15 were differentially expressed in EβF treatment vs. CK and EβF treatment vs. SC. In total, 28 juvenile hormone-related genes were differentially expressed (Figure 2).
FIGURE 2.
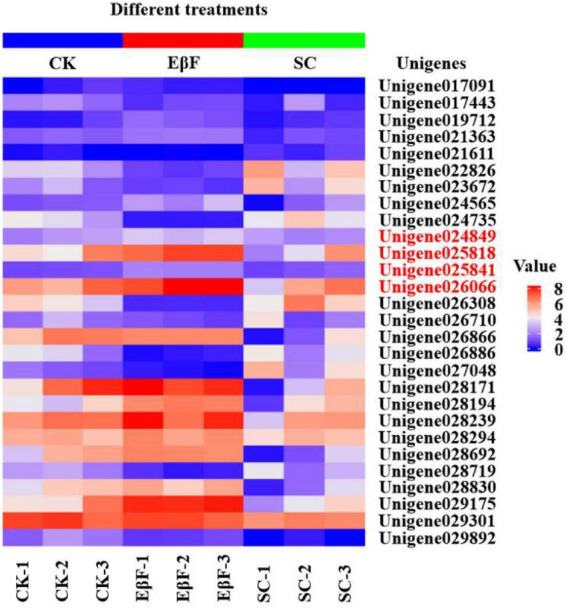
The heat map showed the relative expression level of all differentially expressed juvenile hormone-related unigenes in S. exigua larvae after EβF treatments at doses of 4 g/kg. CK (blank control) and SC (solvent control) were set as controls. The concentrations of EβF treatments were set as 4 g/kg; the units of EβF treatment as g/kg in all treatment groups mean the mass ratio of EβF/artificial diet. The value denotes the value of log2 (FPKM + 1). The unigenes for further study using the RT-qPCR were marked with red font.
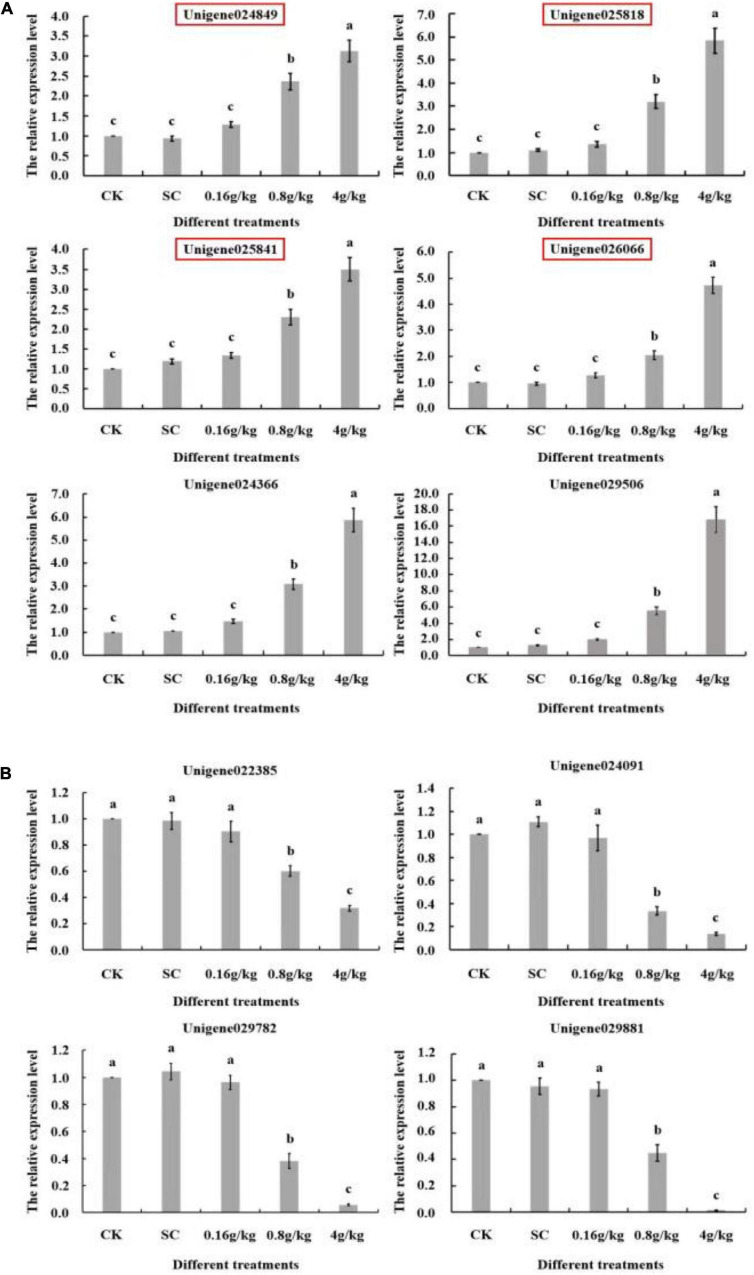
To verify the differential RNA-seq results of S. exigua treated by EβF at a dose of 4 g/kg, 10 genes of S. exigua were selected to be performing RT-qPCR based on the annotation information, including six upregulated and four downregulated genes. Among these ten genes, four important juvenile hormone-related genes were selected and the other six were related to insect cuticle protein, fatty acid metabolism, key enzymes of detoxification metabolism, etc. The detailed information on the selected 10 unigenes is listed in Supplementary Tables 1–3, (Figures 3A,B). These results not only proved the accuracy of differential RNA-seq data but also showed that EβF disrupted the metabolism of the juvenile hormone of S. exigua. Thus, EβF might be used as an antagonist for disrupting the hormone metabolism of S. exigua.
FIGURE 3.
The relative expression of the identified random ten unigenes explored by differential RNA-seq in S. exigua larvae after EβF treatments at different doses. (A) The relative expression of six upregulated unigenes in S. exigua after EβF treatments at different doses. (B) The relative expression of four downregulated unigenes in S. exigua after EβF treatments at different doses. CK (blank control) and SC (solvent control) were set as controls. The concentrations of EβF treatments were set as 0.16, 0.8, and 4 g/kg; the units of g/kg in all treatment groups mean the mass ratio of EβF/artificial diet. Different lower-case letters indicated a significant difference determined by performing ANOVA, followed by Tukey’s HSD test (p < 0.05).
Relative expression of SeVg and SeVgR in Spodoptera exigua treated with (E)-β-farnesene at different concentrations
The expression levels of SeVg decreased significantly with the increase in the concentration of EβF from 0 g/kg (CK and SC controls) to 4 g/kg (p < 0.05; Figure 4). After treatment with 4 g/kg of EβF, the expression of SeVg was reduced by about 50% compared to its expression in the control groups. After treatment with 4 g/kg of EβF, the expression of SeVgR showed a similar change as that of SeVg; thus, indicating that EβF affected the formation and accumulation of vitellogenin. Additionally, SeVgR expression decreased significantly after treatment with 0.8 and 4 g/kg of EβF compared to its expression in the controls (p < 0.05; Figure 4). However, the expression of SeVgR showed non-significant change after treatment with 0.16 g/kg of EβF (p > 0.05; Figure 4). The expression of SeVgR after treatment with 0.8 g/kg of EβF decreased by nearly 50% compared to its expression in the controls.
FIGURE 4.
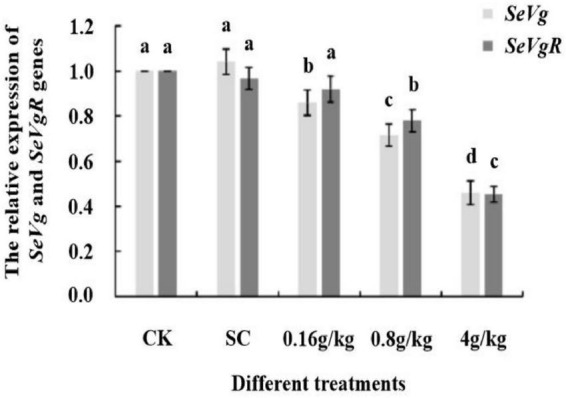
The relative expression of SeVg and SeVgR genes in female adults of S. exigua after EβF treatments at different doses. CK (blank control) and SC (solvent control) were set as controls. The concentrations of EβF treatments were set as 0.16, 0.8, and 4 g/kg; the units of g/kg in all treatment groups means the mass ratio of EβF/artificial diet. Different lower-case letters indicated a significant difference determined by performing ANOVA, followed by Tukey’s HSD test (p < 0.05).
Relative expression of the SeVg and SeVgR proteins in Spodoptera exigua after treatment with different concentrations of (E)-β-farnesene
The results of the Western blot analysis showed that the level of expression of the SeVg and SeVgR proteins was significantly reduced after treatment with 0.16, 0.8, and 4 g/kg of EβF compared to their expression in the two control groups (p < 0.05; Figure 5). The reduction in per female fecundity of S. exigua after treatment with EβF might be due to a decrease in the expression of SeVg and SeVgR, which reduced the intrinsic growth rate and limited the population size of S. exigua.
FIGURE 5.
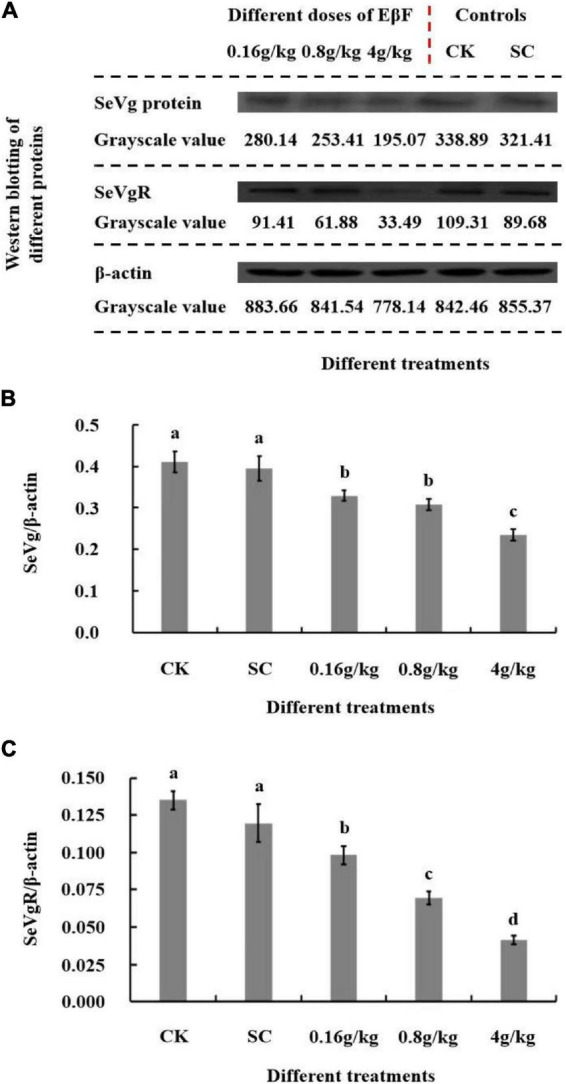
The Western blot assay showed the relative expressions of SeVg and SeVgR proteins in female adults of S. exigua after EβF treatments at different doses. (A) The Western blot pictures of SeVg and SeVgR proteins in female adults of S. exigua after EβF treatments at different doses. (B,C) The relative expressions of SeVg and SeVgR protein expressions by calculating the grayscale value ratios of SeVg/β-actin and SeVgR/β-actin with different treatments. CK (blank control) and SC (solvent control) were set as controls. The concentrations of EβF treatments were set as 0.16, 0.8, and 4 g/kg; the units of g/kg in all treatment groups mean the mass ratio of EβF/artificial diet. Different lower-case letters indicated a significant difference determined by performing ANOVA, followed by Tukey’s HSD test (p < 0.05).
The linear correlation analysis
The linear correlation analysis indicated that the expression of SeVg and SeVgR was significantly positively correlated with per female fecundity (p = 0.0045 and p = 0.0001, respectively; Table 3), hatching rate of eggs (%) in S. exigua (p = 0.067 and p = 0.0003, respectively; Table 3) and the intrinsic growth rate (p = 0.0045 and p = 0.0001, respectively; Table 3). Furthermore, a significant correlation between the hatching rate of eggs (%) and per female fecundity was also found (p = 0.0006, Table 3). In addition, the intrinsic rate of increase was significantly positively correlated with per female fecundity (p = 0.0001; Table 3) and hatching rate of eggs (%) in S. exigua (p = 0.0003; Table 3). Finally, the expression level of SeVg was positively and significantly correlated with the expression level of SeVgR (p = 0.0059, Table 3).
TABLE 3.
The linear correlations between respective key life parameters and SeVgR/SeVg expression in S. exigua after treatment.
| Key life parameters (y) | Expression and other parameters (x) | Linear equation | R 2 | P | F | SE |
| Per female fecundity (y) | The expression of SeVg (x) | y = 614.358x–99.833 | 0.953 | 0.0045* | 60.215 | 37.406 |
| The expression of SeVgR (x) | y = 663.605x–145.490 | 0.999 | 0.0001* | 2507.210 | 5.936 | |
| Hatching rate of eggs (%) (y) | The expression of SeVg (x) | y = 17.216x + 50.663 | 0.938 | 0.0067* | 45.465 | 1.206 |
| The expression of SeVgR (x) | y = 18.671x + 49.322 | 0.992 | 0.0003* | 356.107 | 0.443 | |
| Intrinsic rate of increase (y) | The expression of SeVg (x) | y = 0.129x + 0.025 | 0.952 | 0.0045* | 59.663 | 0.008 |
| The expression of SeVgR (x) | y = 0.139x + 0.016 | 0.998 | 0.0001* | 2300.883 | 0.001 | |
| Per female fecundity (y) | Hatching rate of eggs (%) (x) | y = 35.184x–1875.302 | 0.987 | 0.0006* | 228.106 | 19.564 |
| Intrinsic rate of increase (x) | y = 4773.543x–219.2 | 0.999 | 0.0001* | 2897.987 | 5.522 | |
| Hatching rate of eggs (%) (y) | Intrinsic rate of increase (x) | y = 134.340x + 47.244 | 0.992 | 0.0003* | 385.646 | 0.426 |
| The expression of SeVg (y) | The expression of SeVgR (x) | y = 1.024x–0.028 | 0.943 | 0.0059* | 48.94 | 0.065 |
Data are mean ± standard error; different lower-case letters indicate a significant difference obtained by ANOVA, followed by Tukey’s HSD test (“*” means p < 0.05).
Discussion
Nowadays, exploring natural or synthetic chemicals to kill pests efficiently with the least harm to human or non-target organisms is the focus of studies on pest control (Huang et al., 2021; Tudi et al., 2021). The development of transgenic technology may not only effectively reduce the use of chemical pesticides but also improve the quantity and quality of crops (Kos et al., 2009; Babar et al., 2020). Many transgenic plants are insect resistant, and the most notable one is Bt cotton. Insecticidal toxins from Bacillus thuringiensis (Bt) can provide resistance to the main cotton pest bollworm Helicoverpa armigera (Wu et al., 2008). Additionally, rice containing the Bt gene can produce the protein Cry1Ac, which can effectively control C. suppressalis larvae (Han et al., 2014). However, transgenic technology relies heavily on the discovery of new insect-resistance genes. To achieve a better and wider anti-insect effect, effective insect resistance genes in plants have become an important part of developing transgenic technologies.
The insect resistance gene tps46 was identified in rice (Sun et al., 2017). By overexpressing tps46, eight kinds of metabolites were produced, and the dose ratio of these eight metabolites to the total metabolites was close to 1:104 (Sun et al., 2017). Therefore, standard samples of the secondary metabolites were added to the artificial diet at a ratio of 1:104 to determine the effects of tps46 metabolites on pests, such as C. suppressalis (Yang et al., 2022) and S. exigua (pre-experiment of this study). Our results also indicated that 0.8 g/kg of EβF has a strong inhibitory effect on C. suppressalis larvae, at which concentration of EβF treatment was also reported that the larvae of C. suppressalis were almost all died and were unable to complete their life cycle (Yang et al., 2022). However, at the same concentration of EβF, S. exigua completed its life cycle. Furthermore, 0.8 and 4 g/kg of EβF significantly reduced the survival of S. exigua larvae (p < 0.05), per female fecundity (p < 0.05), and the intrinsic rate of increase value (p < 0.05). Thus, the growth of the population of S. exigua was strongly inhibited. These results suggested that genes, such as tps46 and tps10, which regulate EβF production, might be used as potential resistance genes for pest control (Sun et al., 2017).
After EβF treatment, S. exigua larvae showed abnormal changes, including high mortality of the first and second instar larvae and abnormal pupation of the final mature larvae, compared to the control. Because EβF and juvenile hormone are terpenes and have a similar structure (Noriega, 2014; Li et al., 2021), EβF might affect the normal metabolism of juvenile hormone and disrupt the balance between juvenile hormone and ecdysone, which is essential for normal growth, metamorphosis, and reproduction, eventually causing the death of the S. exigua larvae (Xu et al., 2021). EβF impairs the development and survival of C. suppressalis larvae by disrupting the normal metabolism of juvenile hormone and interfering with the normal hormone balance (Yang et al., 2022). Based on the differential RNA-seq technique, the S. exigua juvenile hormone epoxide hydrolase-related genes (Unigene026066, Unigene025818, and Unigene025841) and the hormone ester-like enzyme-related genes (Unigene024849 and Unigene021363) were found to be significantly upregulated (p < 0.05). Upregulation of these genes increased the degradation of juvenile hormone and disrupted its metabolism (Kamita et al., 2003; El-Sheikh et al., 2016). The details of these genes are presented in Supplementary Table 1. Juvenile hormone is the main gonadotropin of insects. It maintains the larval state and promotes the development of adult ovaries, which is required for reproduction (Gilbert et al., 2000). This could be one of the reasons why some larvae developed late and eventually died, and some successfully developed into adults but per female fecundity was significantly decreased (p < 0.05).
The data from the life table showed that per female fecundity and the intrinsic growth rate of S. exigua after treatment with 0.8 and 4 g/kg of EβF decreased significantly (p < 0.05). These results were similar to those of a study in which 400 ppm of nerolidol significantly inhibited the spawning of Spodoptera littoralis (Ghoneim, 2020), thus indicating that plant secondary metabolites can inhibit the oviposition of pests effectively. Generally known, vitellogenesis is essential for insect reproduction and involves the synthesis of vitellogenin (Vg) and the accumulation of yolk (Xu et al., 2021). In S. exigua, Vg (SeVg) and Vg receptor (SeVgR) are the key genes associated with yolk synthesis and accumulation. Vg is the main nutrient for egg development in pests, and the vitellogenin receptor plays a key role in the uptake of SeVg by oocytes and transport to the ovary (Zhao et al., 2018; Thomas et al., 2020; Hafeez et al., 2021; Wang et al., 2021).
Our results also showed that with an increase in the dose of EβF, the level of expression of both SeVg and SeVgR decreased significantly (p < 0.05). The results of the linear correlation analysis showed that the expression levels of SeVg and SeVgR after treatments with EβF were all significantly related to per female fecundity, the intrinsic growth rate, and the hatching rate of eggs (%) (p < 0.05). All the above findings suggested that EβF should first disturb the normal metabolism of juvenile hormone and break down the balance of juvenile hormone/ecdysone in larvae of S. exigua, and then, it inhibited the expressions of SeVg/SeVgR genes and proteins in female adults of S. exigua. Finally, EβF inhibited the population growth of S. exigua by increasing larval mortality, reducing per female fecundity, and inhibiting the intrinsic growth rate. Therefore, our results in this study indicated that the main plant secondary metabolite, EβF, might be used as a new environmental-friendly insecticidal compound to control the spread of S. exigua in crops.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number: PRJNA764052 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA764052).
Author contributions
YS, H-YD, LY, and Y-JZ conceived and designed the experiments. YS, YL, BJ, WZ, S-MT, and Y-JZ contributed to manuscript writing. YS, YL, WZ, S-MT, BJ, X-TX, Y-XS, LY, and H-YD conducted the experiment. YS, YL, WZ, and S-MT contributed to the data analysis. All authors contributed to the article and approved the submitted version.
Acknowledgments
We are very grateful to Doctor Xiang-dong Kong for his help in the data analysis process of the differentially RNA-seq part and thesis writing. We also thank DBM editing and AIYIDESCI for professional English language editing services.
Footnotes
Funding
This work was supported by the National Natural Science Foundation of China (31871964 and 32100352), the Major Science and Technology Projects in Anhui Province (202003a06020009), the Anhui Provincial Key Laboratory of Molecular Enzymology and Mechanism of Major Diseases in Anhui Normal University (fzmx202007), the Anhui Provincial Natural Science Foundation (Youth project, 2008085QC148), and the Foundation project of Jiangsu Academy of Agricultural Sciences (6111612).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.863626/full#supplementary-material
The annotation of selected ten unigenes.
1,345 differentially expressed genes between EβF vs. CK.
1,345 differentially expressed genes between EβF vs. SC.
References
- Adeyemi M. (2010). The potential of secondary metabolites in plant material as deterents against insect pests: A review. Afr. J. Pure Appl. Chem. 4 243–246. [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq-a Python framework to work with high-throughput sequencing data. Bioinformatics 31 166–169. 10.1093/bioinformatics/btu638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babar U., Nawaz M. A., Arshad U., Azhar M. T., Atif R. M., Golokhvast K. S., et al. (2020). Transgenic crops for the agricultural improvement in Pakistan: A perspective of environmental stresses and the current status of genetically modified crops. GM Crops Food. 11 1–29. 10.1080/21645698.2019.1680078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beale M. H., Birkett M. A., Bruce T. J. A., Chamberlain K., Field L. M., Huttly A. K., et al. (2006). Aphid alarm pheromone produced by transgenic plants affects aphid and parasitoid behavior. Proc. Natl. Acad. Sci. U.S.A. 103 10509–10513. 10.1073/pnas.0603998103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capra E., Colombi C., Depoli P., Nocito F. F., Cocucci M., Vecchietti A., et al. (2015). Protein profiling and tps23 induction in different maize lines in response to methyl jasmonate treatment and Diabrotica virgifera infestation. J. Plant Physiol. 175 68–77. 10.1016/j.jplph.2014.10.018 [DOI] [PubMed] [Google Scholar]
- Chen F., Ro D. K., Petri J., Gershenzon J., Bohlmann J., Pichersky E., et al. (2004). Characterization of a root-specific Arabidopsis terpene synthase responsible for the formation of the volatile monoterpene 1,8-cineole. Plant Physiol. 135 1956–1966. 10.1104/pp.104.044388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Zhou Y., Chen Y. (2018). Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 i884–i890. 10.1093/bioinformatics/bty560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui L. L., Jie D., Francis F., Liu Y. J., Heuskin S., Lognay G., et al. (2012). E-β-farnesene synergizes the influence of an insecticide to improve control of cabbage aphids in China. Crop Prot. 35 91–96. 10.1016/j.cropro.2012.01.003 [DOI] [Google Scholar]
- El-Sheikh S. A., Kamita S. G., Hammock B. D. (2016). Effects of juvenile hormone (JH) analog insecticides on larval development and JH esterase activity in two spodopterans. Pestic. Biochem. Physiol. 128 30–36. 10.1016/j.pestbp.2015.10.008 [DOI] [PubMed] [Google Scholar]
- Fu L., Niu B., Zhu Z., Wu S., Li W. (2012). CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28 3150–3152. 10.1093/bioinformatics/bts565 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoneim K. (2020). Impairing effectiveness of nerolidol, a sesquiterpene compound, on adult performance and reproductive potential of Egyptian cotton leafworm, Spodoptera littoralis (Lepidoptera: Noctuidae). Egypt. Acad. J. Biol. Sci. A Entomol. 13 97–120. 10.21608/EAJBSA.2020.80074 [DOI] [Google Scholar]
- Gilbert L. I., Granger N. A., Roe R. M. (2000). The juvenile hormones: Historical facts and speculations on future research directions. Insect Biochem. Mol. Biol. 30 617–644. 10.1016/S0965-1748(00)00034-5 [DOI] [PubMed] [Google Scholar]
- Greenberg S. M., Sappington T. W., Legaspi B. C., Liu T., Sétamou M. (2001). Feeding and life history of Spodoptera exigua (lepidoptera: Noctuidae) on different host plants. Ann. Entomol. Soc. Am. 94 566–575. 10.1603/0013-87462001094[0566:FALHOS]2.0.CO;2 [DOI] [Google Scholar]
- Gu Z., Elis R., Schlesner M. (2016). Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics 32 2847–2849. 10.1093/bioinformatics/btw313 [DOI] [PubMed] [Google Scholar]
- Hafeez M., Li X., Yousaf H. K., Khan M. M., Imran M., Zhang Z., et al. (2021). Sublethal effects of bistrifluron on key biological traits, macronutrients contents and vitellogenin (SeVg) expression in Spodoptera exigua (Hübner). Pestic. Biochem. Physiol. 174:104802. 10.1016/j.pestbp.2021.104802 [DOI] [PubMed] [Google Scholar]
- Hafeez M., Qasim M., Ali S., Yousaf H. K., Waqas M., Ali E., et al. (2020). Expression and functional analysis of P450 gene induced tolerance/resistance to lambda-cyhalothrin in quercetin fed larvae of beet armyworm Spodoptera exigua (Hübner). Saudi J. Biol. Sci. 27 77–87. 10.1016/j.sjbs.2019.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L., Han C., Liu Z., Chen F., Jurat-Fuentes J. L., Hou M., et al. (2014). Binding site concentration explains the differential susceptibility of Chilo suppressalis and Sesamia inferens to Cry1A-producing rice. Appl. Environ. Microbiol. 80 5134–5140. 10.1128/aem.01544-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L. F., Zhang K. D., Wu Z. W., Xu J. M., Erb M. (2021). Plant volatiles as regulators of plant defense and herbivore immunity: Molecular mechanisms and unanswered questions. Insect Sci. 44 82–88. 10.1016/j.cois.2021.03.010 [DOI] [PubMed] [Google Scholar]
- Huang G., Deng Y., Zhang Y., Feng P., Xu C., Fu L., et al. (2021). Study on long-term pest control and stability of double-layer pesticide carrier in indoor and outdoor environment. Chem. Eng. J. 403:126342. 10.1016/j.cej.2020.126342 [DOI] [Google Scholar]
- Huber D. P. W., Philippe R. N., Godard K. A., Sturrock R. N., Bohlmann J. (2005). Characterization of four terpene synthase cDNAs from methyl jasmonate-induced Douglas-fir, Pseudotsuga menziesii. Phytochemistry 66 1427–1439. 10.1016/j.phytochem.2005.04.030 [DOI] [PubMed] [Google Scholar]
- Kamita S. G., Hinton A. C., Wheelock C. E., Wogulis M., Wilson D. K., Wolf N. M., et al. (2003). Juvenile hormone (JH) esterase: Why are you so JH specific? Insect Biochem. Mol. Biol. 33 1261–1273. 10.1016/j.ibmb.2003.08.004 [DOI] [PubMed] [Google Scholar]
- Kos M., Loon J., Dicke M., Vet L. (2009). Transgenic plants as vital components of integrated pest management. Trends Biotechnol. 27 621–627. 10.1016/j.tibtech.2009.08.002 [DOI] [PubMed] [Google Scholar]
- Langmead B., Trapnell C., Pop M., Salzberg S. L. (2009). Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 10:R25. 10.1186/gb-2009-10-3-r25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Dewey C. N. (2011). RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. 10.1186/1471-2105-12-323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Hu H., Chen Y., Xie J., Li J., Zeng T., et al. (2021). Tissue specificity of (E)-beta-farnesene and germacrene D accumulation in pyrethrum flowers. Phytochemistry 187:112768. 10.1016/j.phytochem.2021.112768 [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods 25 402–408. 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. 10.1186/s13059-014-0550-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michael R., Ranjan A., Kumar R. S., Pathak P. K., Trivedi P. K. (2020). Light-regulated expression of terpene synthase gene, AtTPS03, is controlled by the bZIP transcription factor, HY5, in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 529 437–443. 10.1016/j.bbrc.2020.05.222 [DOI] [PubMed] [Google Scholar]
- Morse A., Kevan P., Shipp L., Khosla S., McGarvey B. (2012). The impact of greenhouse tomato (Solanales: Solanaceae) floral volatiles on bumble bee (Hymenoptera: Apidae) pollination. Environ. Entomol. 41 855–864. 10.1603/EN11234 33044624 [DOI] [Google Scholar]
- Nagegowda D. A., Gupta P. (2020). Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 294:110457. 10.1016/j.plantsci.2020.110457 [DOI] [PubMed] [Google Scholar]
- Noriega F. G. (2014). Juvenile hormone biosynthesis in insects: What is new, what do we know, and what questions remain? Int. Sch. Res. Notices 2014:967361. 10.1155/2014/967361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X. L., Chen R. R., Zhang Y., Ma Y., Cui J. J., Han Z. J., et al. (2013). A Spodoptera exigua cadherin serves as a putative receptor for Bacillus thuringiensis Cry1Ca toxin and shows differential enhancement of Cry1Ca and Cry1Ac toxicity. Appl. Environ. Microbiol. 79 5576–5583. 10.1128/AEM.01519-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saeed Q., Ahmad F., Iqbal N., Zaka S. M. (2019). Chemical control of polyphagous pests on their auxiliary hosts can minimize insecticide resistance: A case study of Spodoptera exigua Hübner (Lepidoptera: Noctuidae) in cotton agroecosystem. Ecotoxicol. Environ. Saf. 171 721–727. 10.1016/j.ecoenv.2019.01.038 [DOI] [PubMed] [Google Scholar]
- Saeed S., Sayyed A. H., Ahmad I. (2010). Effect of host plants on life-history traits of Spodoptera exigua (Lepidoptera: Noctuidae). J. Pest Sci. 83 165–172. 10.1007/s10340-009-0283-8 [DOI] [Google Scholar]
- Schnee C., Kollner T. G., Held M., Turlings T. C. J., Gershenzon J., Degenhardt J. (2006). The products of a single maize sesquiterpene synthase form a volatile defense signal that attracts natural enemies of maize herbivores. Proc. Natl. Acad. Sci. U.S.A. 103 1129–1134. 10.1073/pnas.0508027103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song N. N., Ding W. H., Chu S. Y., Zhao J., Dong X., Di B. B., et al. (2012). Urotensin II stimulates vascular endothelial growth factor secretion from adventitial fibroblasts in synergy with Angiotensin II. Vascular Med. 76 1267–1273. 10.1253/circj.CJ-11-0870 [DOI] [PubMed] [Google Scholar]
- Su S., Liu X., Pan G., Hou X., Yuan Y. (2015). In vitro characterization of a (E)-β-farnesene synthase from Matricaria recutita L. and its up-regulation by methyl jasmonate. Gene 571 58–64. 10.1016/j.gene.2015.06.037 [DOI] [PubMed] [Google Scholar]
- Sun Y., Huang X., Ning Y., Jing W., Bruce T. J. A., Qi F., et al. (2017). TPS46, a rice terpene synthase conferring natural resistance to bird cherry-oat aphid, Rhopalosiphum padi (Linnaeus). Front. Plant Sci. 8:110. 10.3389/fpls.2017.00110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Xiao L., Cao G., Zhang Y., Xiao Y., Xu G., et al. (2016). Molecular characterisation of the vitellogenin gene (AlVg) and its expression after Apolygus lucorum had fed on different hosts. Pest Manag. Sci. 72 1743–1751. 10.1002/ps.4203 [DOI] [PubMed] [Google Scholar]
- Thomas D. S., Varma M., Manoharan C., Rao G. P., Vijayan K., Mishra R. K., et al. (2020). Molecular characterization and functional analysis of the vitellogenin receptor from eri silkworm, Samia ricini. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 242:110417. 10.1016/j.cbpb.2020.110417 [DOI] [PubMed] [Google Scholar]
- Tudi M., Ruan H. D., Wang L., Lyu J., Sadler R., Connell D., et al. (2021). Agriculture development, pesticide application and its impact on the environment. Int. J. Environ. Res. Public Health 18:1112. 10.3390/ijerph18031112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B., Dong W., Li H., D’Onofrio C., Bai P., Chen R., et al. (2022). Molecular basis of (E)-beta-farnesene-mediated aphid location in the predator Eupeodes corollae. Curr. Biol. 32 951-962.e7. 10.1016/j.cub.2021.12.054 [DOI] [PubMed] [Google Scholar]
- Wang S., Miao S., Yang B., Wang Z., Liu Q., Wang R., et al. (2021). Initial characterization of the vitellogenin receptor from a Psocoptera insect: Function analysis and RNA interference in Liposcelis entomophila (Enderlein). J. Stored Products Res. 92:101803. 10.1016/j.jspr.2021.101803 [DOI] [Google Scholar]
- War A. R., Paulraj M. G., Ahmad T., Buhroo A. A., Hussain B., Ignacimuthu S., et al. (2012). Mechanisms of plant defense against insect herbivores. Plant Signal. Behav. 7 1306–1320. 10.4161/psb.21663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu K. M., Lu Y. H., Feng H. Q., Jiang Y. Y., Zhao J. Z. (2008). Suppression of cotton bollworm in multiple crops in China in areas with Bt toxin-containing cotton. Science 321 1676–1678. 10.1126/science.1160550 [DOI] [PubMed] [Google Scholar]
- Xu H., Zhang Y., Zhang L., Wang Z., Guo P., Zhao P. (2021). Structural characterization and functional analysis of juvenile hormone diol kinase from the silkworm, Bombyx mori. Int. J. Biol. Macromol. 167 570–577. 10.1016/j.ijbiomac.2020.11.138 [DOI] [PubMed] [Google Scholar]
- Yang L., Yao X. M., Liu B. S., Han Y. C., Ji R., Ju J. F., et al. (2022). Caterpillar-induced rice volatile (E)-β-Farnesene impairs the development and survival of Chilo suppressalis larvae by disrupting insect hormone balance. Front. Physiol. 13:904482. 10.3389/fphys.2022.904482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X., Zhang Y., Ma Y., Xu Z., Wang G., Xia L. (2013). Expression of an (E)-β-farnesene synthase gene from Asian peppermint in tobacco affected aphid infestation. Crop J. 1 50–60. 10.1016/j.cj.2013.07.005 [DOI] [Google Scholar]
- Yuan J. S., Kollner T. G., Wiggins G., Grant J., Degenhardt J., Chen F. (2008). Molecular and genomic basis of volatile-mediated indirect defense against insects in rice. Plant J. Cell Mol. Biol. 55 491–503. 10.1111/j.1365-313X.2008.03524.x [DOI] [PubMed] [Google Scholar]
- Zhang B., Liu H., Hull-Sanders H., Wang J. (2011). Effect of host plants on development, fecundity and enzyme activity of Spodoptera exigua (Hübner) (Lepidoptera: Noctuidae). Agricult. Sci. China 10 1232–1240. 10.1016/S1671-2927(11)60114-4 [DOI] [Google Scholar]
- Zhao J., Sun Y., Xiao L., Tan Y., Jiang Y., Bai L. (2018). Vitellogenin and vitellogenin receptor gene expression profiles in Spodoptera exigua are related to host plant suitability. Pest Manag. Sci. 74 950–958. 10.1002/ps.4794 [DOI] [PubMed] [Google Scholar]
- Zhen C., Gao X. (2016). A point mutation (L1015F) of the voltage-sensitive sodium channel gene associated with lambda-cyhalothrin resistance in Apolygus lucorum (Meyer-Dür) population from the transgenic Bt cotton field of China. Pestic. Biochem. Physiol. 127 82–89. 10.1016/j.pestbp.2015.09.011 [DOI] [PubMed] [Google Scholar]
- Zhou F., Pichersky E. (2020). More is better: The diversity of terpene metabolism in plants. Curr. Opin. Plant Biol. 55 1–10. 10.1016/j.pbi.2020.01.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The annotation of selected ten unigenes.
1,345 differentially expressed genes between EβF vs. CK.
1,345 differentially expressed genes between EβF vs. SC.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: National Center for Biotechnology Information (NCBI) BioProject database under accession number: PRJNA764052 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA764052).